Gene
KWMTBOMO12226
Annotation
endonuclease-reverse_transcriptase_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.558
Sequence
CDS
ATGTGGATGAATCTGGAAAGTAAAATAAATGATGCCCTTGTAGATGTTACCTCTCAAAGAGAAAAAGATCCCAGGAGATCCACATGGATAACAGACCAAACATGGAGCCTAATCCAGCGCAGAAAAGATGTCAAAGTCAGAGGCCTACAGAATCCTGAGATACGTAGGGAATACTCTGAGATTTGTAAAGCAGTCCAAAGAGAATGCCGCAGAGACAAAAATAAATTTATTATGGGCATATGTCGAGAGGTGGAATCACACGCGCTAAAGCTGCAAACTTCAGATCTGTTCAAGAAAGTCCGTCTGCTAAGTAGAAAATTCAAGCCCAAATCTTGGGTCATAGAAAACTCTGAAGGACTACCACTGCATGAACTGCACGAAATCACGGAACGCTGGCGTGGCTACTGTGAGCAACTCTATAAAGCTCCCCATGAATCTACCGAAAGCCGGTTCGACTGGAAAGATCTTGAATTGGAGCCTAATATTCTGAGATCAGAGGTAACGGGTGCAATCGACCGGCTCAAATCAAAGAAGGCTCCCGGACCTGACGATATCACATCGGAGATCCTCAAGGCTCTTGGTGACAATGGTTTGGATGTACTCCATAAGATTTGCAATCACGTATGGCAAAACGGTGAATGGCCCAAAAACTGGACGCGGTCTATAATTATCCCACTGCACAAGAAAGGTTCAACTAGAAAATGTGATAATTACCGCACCCTGGCCCTCATCTCTCACGCTAGCAAAGTTCTTTTGCATGTAATAAACAGCAGGATTCGTTACTATTTGGACTGGCAGATCCCGCAGGAACAAGCTGGATTTGTAAAAGGCAAGGGCACGAGGGAGCAAATACTGAACGTGAGACAGATTGTGGAAAAGTGCTACGAATTCGACACGCCGGTCATCATGTGTTTCGTTGACTATAGTAAGGCTTTTGACTGTGTTGACTGGAACTGCTTGTGGGATGTCCTCACAGAGTTGGGGGTCCCTCGCCACCTTACTGCACTCCTTCAGTCATTATACTACAACAGCCAGGGAGTAGTGAGAATCGATAGGACTGCATCTGACTCGTTCAAATTTCACAAGGGCGTGCGGCAGGGCTGCATCCTATCACCCATCCTGTTCAATATTTACGGCGAATACATCGTCAGAAAAACATGCGATGGATGGGATGGTGGTATTACCGTCGGAGGCTGTAAAATCTCTAACTTACGCTACGCTGACGACACCACTCTCCTGGCAGCCAGCGAAATCGAGATGAGCAAGTTCTTGGACAGGATGGAGCGCATTAGCAAAGACCTCGGACTCGTGATAAACAGATCTAAAACAAAGGTCATGATCGTTGATCGCACAGGCAGACTGGAGCTCAGCGGTACACTTAATTTAGAAGTAACGGACAACTTTGTTTACCTGGGCTCCAACATCAGCAACAATGGCTCTTGTGAACCGGAAATTCGTAGACGGATTGGGATGGCAAAAGGTGCCATGTCTCAGCTGGACAAAATATGGAAGGACAAAAATATCTCTATGAACACCAAGAGCAACCTAGTCCGCACACTAGTGTTTTCAATATTCATATACGGAGCAGAGACATGGACTATAAAGAAAGCTGACCGCAATCGGATTGATGCCTTCGAAATGTGGTGCTGGAGGAGGATGTTGGGTATTCCCTGGACCGCACGCCGCACCAATTTATCCATCCTAGCTCAGTTGAAGATCCGGACTAGACTCTCCACCATTTGCATGCGAAGGATTCTTGAATACTTTGGCCACATCGCAAGGAAAGACAGTGACAACTTAGAGAAATTGATGGTCACTGGAAAGGTCGAGGGCAAGCGACTAAGAGGCAGGAACCCGACCCGCTGGACGGACCAGATTCGTGCAGCCCTGGATACCACAGTGCACGATGCTCTGCAATCAGCTGCAGATAGGAACAGATGGCGATGCACTATAAAAAACAATCTGTTTCAAGGCCATGACCACGACCCTCAGTAA
Protein
MWMNLESKINDALVDVTSQREKDPRRSTWITDQTWSLIQRRKDVKVRGLQNPEIRREYSEICKAVQRECRRDKNKFIMGICREVESHALKLQTSDLFKKVRLLSRKFKPKSWVIENSEGLPLHELHEITERWRGYCEQLYKAPHESTESRFDWKDLELEPNILRSEVTGAIDRLKSKKAPGPDDITSEILKALGDNGLDVLHKICNHVWQNGEWPKNWTRSIIIPLHKKGSTRKCDNYRTLALISHASKVLLHVINSRIRYYLDWQIPQEQAGFVKGKGTREQILNVRQIVEKCYEFDTPVIMCFVDYSKAFDCVDWNCLWDVLTELGVPRHLTALLQSLYYNSQGVVRIDRTASDSFKFHKGVRQGCILSPILFNIYGEYIVRKTCDGWDGGITVGGCKISNLRYADDTTLLAASEIEMSKFLDRMERISKDLGLVINRSKTKVMIVDRTGRLELSGTLNLEVTDNFVYLGSNISNNGSCEPEIRRRIGMAKGAMSQLDKIWKDKNISMNTKSNLVRTLVFSIFIYGAETWTIKKADRNRIDAFEMWCWRRMLGIPWTARRTNLSILAQLKIRTRLSTICMRRILEYFGHIARKDSDNLEKLMVTGKVEGKRLRGRNPTRWTDQIRAALDTTVHDALQSAADRNRWRCTIKNNLFQGHDHDPQ
Summary
Uniprot
D7F163
D7F168
D7F172
D7F161
D7F162
D5LB39
+ More
D7F175 D7F167 A0A2W1BNH4 A0A2W1BGD9 A0A2H6KKM9 A0A3Q1LQN1 A0A3Q1MLZ9 A0A3Q1MJK8 A0A3Q1M6J7 A0A3S3P648 A0A3Q1MGH6 A0A3Q1MQ02 A0A3Q1N3Y9 A0A3Q1LXQ5 A0A3Q1MMW6 A0A3Q1M1T7 A0A3Q1MP18 A0A3Q1LUA6 A0A3Q1MI63 A0A3Q1LI58 A0A3Q1LWU3 A0A3Q1M527 A0A3Q1LNZ2 A0A3Q1LR18 A0A3Q1M8D1 A0A3Q1N2X2 H3B2Y3 A0A3Q1LXF5 A0A3Q1MWB3 A0A3Q1MNB9 A0A3Q1NE48 A0A3Q1N1P1 A0A3Q1MDY8 A0A3Q1LYT3 A0A3Q1MA23 A0A3Q1MLV4 A0A3Q1MLI2 A0A3Q1ML42 A0A3Q1MZJ0 A0A3Q1LRD2 A0A3Q1LWH0 A0A3Q1MM54 A0A3Q1M0W5 A0A3Q1MXT5 A0A3Q1LR52 A0A3Q1LX64 A0A0J7KGF7 F6RFH2 H2YWZ5 A0A3Q1LIW6 A0A3Q1MKR5 A0A3Q1MGI7 A0A3Q1M8I7 A0A3Q1NFT8 A0A3Q1MEQ0 A0A3Q1NI39 A0A3Q1N0X3 A0A3Q1M6V6 A0A3Q1MLF3 A0A3Q1M4W4 A0A3Q1N0I8 A0A3Q1LWK2 W5N9W4
D7F175 D7F167 A0A2W1BNH4 A0A2W1BGD9 A0A2H6KKM9 A0A3Q1LQN1 A0A3Q1MLZ9 A0A3Q1MJK8 A0A3Q1M6J7 A0A3S3P648 A0A3Q1MGH6 A0A3Q1MQ02 A0A3Q1N3Y9 A0A3Q1LXQ5 A0A3Q1MMW6 A0A3Q1M1T7 A0A3Q1MP18 A0A3Q1LUA6 A0A3Q1MI63 A0A3Q1LI58 A0A3Q1LWU3 A0A3Q1M527 A0A3Q1LNZ2 A0A3Q1LR18 A0A3Q1M8D1 A0A3Q1N2X2 H3B2Y3 A0A3Q1LXF5 A0A3Q1MWB3 A0A3Q1MNB9 A0A3Q1NE48 A0A3Q1N1P1 A0A3Q1MDY8 A0A3Q1LYT3 A0A3Q1MA23 A0A3Q1MLV4 A0A3Q1MLI2 A0A3Q1ML42 A0A3Q1MZJ0 A0A3Q1LRD2 A0A3Q1LWH0 A0A3Q1MM54 A0A3Q1M0W5 A0A3Q1MXT5 A0A3Q1LR52 A0A3Q1LX64 A0A0J7KGF7 F6RFH2 H2YWZ5 A0A3Q1LIW6 A0A3Q1MKR5 A0A3Q1MGI7 A0A3Q1M8I7 A0A3Q1NFT8 A0A3Q1MEQ0 A0A3Q1NI39 A0A3Q1N0X3 A0A3Q1M6V6 A0A3Q1MLF3 A0A3Q1M4W4 A0A3Q1N0I8 A0A3Q1LWK2 W5N9W4
EMBL
FJ265548
ADI61816.1
FJ265553
ADI61821.1
FJ265557
ADI61825.1
+ More
FJ265546 ADI61814.1 FJ265547 ADI61815.1 GU815090 ADF18553.1 FJ265560 ADI61828.1 FJ265552 ADI61820.1 KZ150025 PZC74827.1 KZ150203 PZC72267.1 BDSA01000081 GBE63537.1 NCKU01004395 RWS06003.1 AFYH01119920 LBMM01007694 KMQ89503.1 AHAT01022314
FJ265546 ADI61814.1 FJ265547 ADI61815.1 GU815090 ADF18553.1 FJ265560 ADI61828.1 FJ265552 ADI61820.1 KZ150025 PZC74827.1 KZ150203 PZC72267.1 BDSA01000081 GBE63537.1 NCKU01004395 RWS06003.1 AFYH01119920 LBMM01007694 KMQ89503.1 AHAT01022314
Proteomes
Interpro
SUPFAM
SSF56219
SSF56219
Gene 3D
ProteinModelPortal
D7F163
D7F168
D7F172
D7F161
D7F162
D5LB39
+ More
D7F175 D7F167 A0A2W1BNH4 A0A2W1BGD9 A0A2H6KKM9 A0A3Q1LQN1 A0A3Q1MLZ9 A0A3Q1MJK8 A0A3Q1M6J7 A0A3S3P648 A0A3Q1MGH6 A0A3Q1MQ02 A0A3Q1N3Y9 A0A3Q1LXQ5 A0A3Q1MMW6 A0A3Q1M1T7 A0A3Q1MP18 A0A3Q1LUA6 A0A3Q1MI63 A0A3Q1LI58 A0A3Q1LWU3 A0A3Q1M527 A0A3Q1LNZ2 A0A3Q1LR18 A0A3Q1M8D1 A0A3Q1N2X2 H3B2Y3 A0A3Q1LXF5 A0A3Q1MWB3 A0A3Q1MNB9 A0A3Q1NE48 A0A3Q1N1P1 A0A3Q1MDY8 A0A3Q1LYT3 A0A3Q1MA23 A0A3Q1MLV4 A0A3Q1MLI2 A0A3Q1ML42 A0A3Q1MZJ0 A0A3Q1LRD2 A0A3Q1LWH0 A0A3Q1MM54 A0A3Q1M0W5 A0A3Q1MXT5 A0A3Q1LR52 A0A3Q1LX64 A0A0J7KGF7 F6RFH2 H2YWZ5 A0A3Q1LIW6 A0A3Q1MKR5 A0A3Q1MGI7 A0A3Q1M8I7 A0A3Q1NFT8 A0A3Q1MEQ0 A0A3Q1NI39 A0A3Q1N0X3 A0A3Q1M6V6 A0A3Q1MLF3 A0A3Q1M4W4 A0A3Q1N0I8 A0A3Q1LWK2 W5N9W4
D7F175 D7F167 A0A2W1BNH4 A0A2W1BGD9 A0A2H6KKM9 A0A3Q1LQN1 A0A3Q1MLZ9 A0A3Q1MJK8 A0A3Q1M6J7 A0A3S3P648 A0A3Q1MGH6 A0A3Q1MQ02 A0A3Q1N3Y9 A0A3Q1LXQ5 A0A3Q1MMW6 A0A3Q1M1T7 A0A3Q1MP18 A0A3Q1LUA6 A0A3Q1MI63 A0A3Q1LI58 A0A3Q1LWU3 A0A3Q1M527 A0A3Q1LNZ2 A0A3Q1LR18 A0A3Q1M8D1 A0A3Q1N2X2 H3B2Y3 A0A3Q1LXF5 A0A3Q1MWB3 A0A3Q1MNB9 A0A3Q1NE48 A0A3Q1N1P1 A0A3Q1MDY8 A0A3Q1LYT3 A0A3Q1MA23 A0A3Q1MLV4 A0A3Q1MLI2 A0A3Q1ML42 A0A3Q1MZJ0 A0A3Q1LRD2 A0A3Q1LWH0 A0A3Q1MM54 A0A3Q1M0W5 A0A3Q1MXT5 A0A3Q1LR52 A0A3Q1LX64 A0A0J7KGF7 F6RFH2 H2YWZ5 A0A3Q1LIW6 A0A3Q1MKR5 A0A3Q1MGI7 A0A3Q1M8I7 A0A3Q1NFT8 A0A3Q1MEQ0 A0A3Q1NI39 A0A3Q1N0X3 A0A3Q1M6V6 A0A3Q1MLF3 A0A3Q1M4W4 A0A3Q1N0I8 A0A3Q1LWK2 W5N9W4
PDB
5HHL
E-value=0.0141258,
Score=92
Ontologies
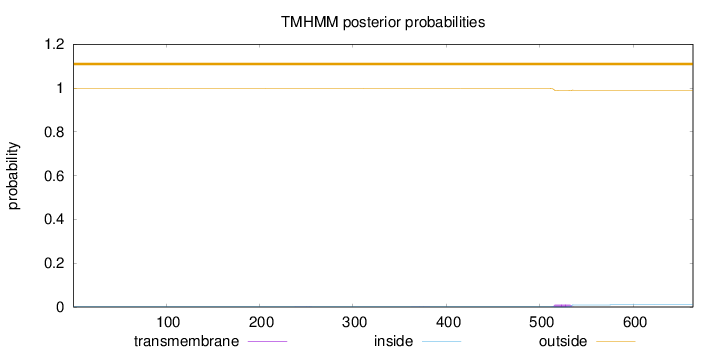
Topology
Length:
664
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.22052
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00233
outside
1 - 664
Population Genetic Test Statistics
Pi
213.404976
Theta
56.146862
Tajima's D
1.636366
CLR
0.016504
CSRT
0.822908854557272
Interpretation
Uncertain
