Gene
KWMTBOMO12225 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA014145
Annotation
PREDICTED:_myrosinase_1-like_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.314
Sequence
CDS
ATGGTTCAAGTTCGGGGCGGCCACGTCGTCGTACCAGATCGAAGGGTCCTGGAATACAAAGCAATAAGTATTTGGGATGATTACACGCACACACAGCCCCATAAGATCAAAGATCACTCCAACGGAGATGTTGCTTGTGACTCTTACAACCAGTACGAGAAGGACATTGAGATGGCCGAGCAGTTAGGACTTGATTTTTACAGGTTTTCTATATCCTGGTCAAGACTGCTGCCTGAGGGATACCGCACTCGCATCAGTTCGGACGGACGGCAATACTACGACAATTTGATCAACGGACTGCTAGACAGAGGGATTGAACCTATGGTCACTTTGTACCATTCAGAAATGCCGAAGATGTTTCAAGATTTGGGAGGCTGGGCCAATCCTGAAATGGCAGAATGGTTTGCCGATTACGCGGATGTAGCATTCTCTCTGTACGGAGATAGAGTGAAGCTTTGGATCACGATCAATGAGCCTTCAATTATTTGTGATAGCAGTGCATACGTTCTAGGAGTCGTCGAGCCATTCCTGGCTCCATATTTGTGCTCGAAGACTGTACTTATGGCTCATGCCAAAGCTTACCATAATTATAAAAAGAACTATAAGCCGATGTATCAAGGGAAAGTTGGTATCGCCCACGCCGCATCTTATTTCAGTGCAAAAACTCAAAACGATGCGGAGCTTGCAGAACTATTCCGGCAATATCATTTTGGTCTTTTTATGCATCCATTACATTCCAAGTCTGGAGGGTGGCCTGCAACCGTTGTAGAACTCATGAAGAATATCAGCTCAGTCGAAGGATTCCCGCGCTCTAGATTACCTCCGTTAACCAAACATGAAGTTGAATTCATCAGAGGTACTAACGATTTCCTTGGACTTAACTACTACACCAGCTGCACGGTGAGAAAAGCTAAAGCTGGCGAGAAAAGGCAGGAATCCCTTGGTGATCTGCCAGAGTTGAACGCGATTTTCGAACCTTACCCTAACAGCACTGCGGCATATCCTGAATATTACAGCTCATACCCTGAAGGCCTGAGGGCGTTCATAGCTTGGTTCACGAAAAATTACGGAGTCCAAGATATTTTCATCACAGAAAACGGTTACACCAGTTACACAGATGACCTGAACGATTTCAACAGAATCACTTACATTGAGAAACATTTGGAGAAGATTCTACTTTGCATTTACGAAGACGGAGCTAGAATACACGGTTACACTTACTGGTCACTGATGGACAACTTCGAATGGCGAAACGGGTACACAGCAAAGTTTGGTCTTTACGACGTCGACTTCTCTGACGAGGGCCGGAAGCGCACTCCAAGACTCTCAGCCCAGTACTACTCATACGTTATCAAGAGCAGATCGATCGATTCTAACAAGTGGAAGTCACAGATGGAAGATACGAACGACAACTAA
Protein
MVQVRGGHVVVPDRRVLEYKAISIWDDYTHTQPHKIKDHSNGDVACDSYNQYEKDIEMAEQLGLDFYRFSISWSRLLPEGYRTRISSDGRQYYDNLINGLLDRGIEPMVTLYHSEMPKMFQDLGGWANPEMAEWFADYADVAFSLYGDRVKLWITINEPSIICDSSAYVLGVVEPFLAPYLCSKTVLMAHAKAYHNYKKNYKPMYQGKVGIAHAASYFSAKTQNDAELAELFRQYHFGLFMHPLHSKSGGWPATVVELMKNISSVEGFPRSRLPPLTKHEVEFIRGTNDFLGLNYYTSCTVRKAKAGEKRQESLGDLPELNAIFEPYPNSTAAYPEYYSSYPEGLRAFIAWFTKNYGVQDIFITENGYTSYTDDLNDFNRITYIEKHLEKILLCIYEDGARIHGYTYWSLMDNFEWRNGYTAKFGLYDVDFSDEGRKRTPRLSAQYYSYVIKSRSIDSNKWKSQMEDTNDN
Summary
Similarity
Belongs to the glycosyl hydrolase 1 family.
Uniprot
H9JX80
H9JX83
H9JXC4
A0A3S2N597
A0A2A4J905
A0A2A4IWH6
+ More
A0A2W1BMB0 A0A194QH27 A0A2W1BC39 H9JX79 A0A2H1W6N8 A0A3G1T1E9 A0A2H1WLJ0 A0A2H1VC49 A0A194QFC2 A0A194QFH9 A0A2A4IZ96 A0A2W1BEU6 H9JXC5 A0A2W1BJB1 H9JXC7 A0A0T6B9C6 A0A0K8UIW8 A0A034VAL4 A0A0K8W4X1 A0A034VEY7 A0A0K8W3Q7 A0A1W4XQQ3 Q7Q5I1 A0A0A1XAG3 D6X3T6 A0A212FF93 A0A1Y9IS68 A0A212FF90 A0A194PJU3 A0A1Y9GLV6 G9F9I3 W8BL15 A0A194RQF8 A0A0A1XS57 N6TKX9 J3JV61 A0A212EXF5 A0A3S2PE71 A0A194PMT2 A0A1Y9J0B2 A0A1A9W4X6 A0A182JGM2 A0A182LL18 A0A1B0C2P0 D0VYR9 A0A1A9XR01 Q7PI98 A0A194R8P7 A0A2W1BWF3 D0VYS0 A0A1A9UP13 A0A194PPG4 A0A1Y9GL87 A0A1A9ZLX0 A0A182TBP9 A0A1Y9IW73 A0A1Y9J0J6 D0VYS1 A0A182YP84 A0A1Y1K6D9 A0A194R3H6 A0A182TWE4 B2DBM6 A0A2A4JPS5 A0A1Y9IS75 A0A2J7Q530 A0A182U0B4 Q5TQY8 A0A1Y9GKT0 A0A1Y9IRY8 D0VYR6 A0A2H1VF65 A0A194R3H3 G9F9G9 A0A194R360 D5KQK3 A0A194R3Z4 B0WNN7 D0VYR5 A0A1S5SJI5 A0A219WGQ6 X2CKZ4 W5JK33 A0A194QYR8 A0A2A4IZK8 S5SP57 A0A182LWK9 D0VYR8 A0A194PNC9 A0A2M4CLR8 A0A1B6D777 Q16WF2 A0A182YP82 W5JPP3
A0A2W1BMB0 A0A194QH27 A0A2W1BC39 H9JX79 A0A2H1W6N8 A0A3G1T1E9 A0A2H1WLJ0 A0A2H1VC49 A0A194QFC2 A0A194QFH9 A0A2A4IZ96 A0A2W1BEU6 H9JXC5 A0A2W1BJB1 H9JXC7 A0A0T6B9C6 A0A0K8UIW8 A0A034VAL4 A0A0K8W4X1 A0A034VEY7 A0A0K8W3Q7 A0A1W4XQQ3 Q7Q5I1 A0A0A1XAG3 D6X3T6 A0A212FF93 A0A1Y9IS68 A0A212FF90 A0A194PJU3 A0A1Y9GLV6 G9F9I3 W8BL15 A0A194RQF8 A0A0A1XS57 N6TKX9 J3JV61 A0A212EXF5 A0A3S2PE71 A0A194PMT2 A0A1Y9J0B2 A0A1A9W4X6 A0A182JGM2 A0A182LL18 A0A1B0C2P0 D0VYR9 A0A1A9XR01 Q7PI98 A0A194R8P7 A0A2W1BWF3 D0VYS0 A0A1A9UP13 A0A194PPG4 A0A1Y9GL87 A0A1A9ZLX0 A0A182TBP9 A0A1Y9IW73 A0A1Y9J0J6 D0VYS1 A0A182YP84 A0A1Y1K6D9 A0A194R3H6 A0A182TWE4 B2DBM6 A0A2A4JPS5 A0A1Y9IS75 A0A2J7Q530 A0A182U0B4 Q5TQY8 A0A1Y9GKT0 A0A1Y9IRY8 D0VYR6 A0A2H1VF65 A0A194R3H3 G9F9G9 A0A194R360 D5KQK3 A0A194R3Z4 B0WNN7 D0VYR5 A0A1S5SJI5 A0A219WGQ6 X2CKZ4 W5JK33 A0A194QYR8 A0A2A4IZK8 S5SP57 A0A182LWK9 D0VYR8 A0A194PNC9 A0A2M4CLR8 A0A1B6D777 Q16WF2 A0A182YP82 W5JPP3
Pubmed
EMBL
BABH01035816
BABH01035817
BABH01035811
BABH01035812
BABH01035821
RSAL01000405
+ More
RVE41892.1 NWSH01002416 PCG68339.1 NWSH01005512 PCG64155.1 KZ150152 PZC72823.1 KQ459053 KPJ04240.1 PZC72822.1 BABH01035825 ODYU01006661 SOQ48703.1 MG992371 AXY94809.1 ODYU01009444 SOQ53877.1 ODYU01001752 SOQ38385.1 KPJ04243.1 KPJ04242.1 NWSH01004417 PCG65145.1 PZC72831.1 BABH01035826 BABH01035827 PZC72820.1 BABH01035838 BABH01035839 LJIG01003002 KRT83938.1 GDHF01025685 JAI26629.1 GAKP01018616 JAC40336.1 GDHF01006374 JAI45940.1 GAKP01018617 JAC40335.1 GDHF01006779 JAI45535.1 AAAB01008960 EAA11668.3 GBXI01006185 JAD08107.1 KQ971326 EEZ97373.2 AGBW02008847 OWR52397.1 OWR52398.1 KQ459601 KPI93701.1 APCN01002262 JN033728 AEW46881.1 GAMC01007113 JAB99442.1 KQ459875 KPJ19634.1 GBXI01000924 JAD13368.1 APGK01033064 APGK01033065 APGK01033066 KB740860 ENN78593.1 BT127127 AEE62089.1 AGBW02011758 OWR46180.1 RSAL01000077 RVE48784.1 KQ459598 KPI94627.1 JXJN01024604 JXJN01024605 JXJN01024606 AB508958 BAI50022.1 EAA44227.3 KQ460779 KPJ12246.1 KZ149936 PZC77130.1 AB508959 BAI50023.1 KPI94624.1 AB508960 BAI50024.1 GEZM01091306 JAV56934.1 KPJ12247.1 AB264710 BAG30785.1 NWSH01000819 PCG74065.1 NEVH01018372 PNF23684.1 EAL40075.2 AB508955 BAI50019.1 ODYU01001971 SOQ38904.1 KPJ12242.1 JN033714 AEW46867.1 KPJ12243.1 GU591172 ADD92156.1 KPJ12244.1 DS232013 EDS31873.1 AB508954 AB508956 BAI50018.1 BAI50020.1 KU170546 APM84101.1 KT992464 AOY34571.1 KC533771 AGP76179.1 ADMH02000892 ETN64737.1 KQ461150 KPJ08731.1 NWSH01004297 PCG65255.1 KC891004 AGS32242.1 AXCM01009805 AB508957 BAI50021.1 KQ459597 KPI94941.1 GGFL01002037 MBW66215.1 GEDC01015759 JAS21539.1 CH477568 EAT38910.1 ETN64734.1
RVE41892.1 NWSH01002416 PCG68339.1 NWSH01005512 PCG64155.1 KZ150152 PZC72823.1 KQ459053 KPJ04240.1 PZC72822.1 BABH01035825 ODYU01006661 SOQ48703.1 MG992371 AXY94809.1 ODYU01009444 SOQ53877.1 ODYU01001752 SOQ38385.1 KPJ04243.1 KPJ04242.1 NWSH01004417 PCG65145.1 PZC72831.1 BABH01035826 BABH01035827 PZC72820.1 BABH01035838 BABH01035839 LJIG01003002 KRT83938.1 GDHF01025685 JAI26629.1 GAKP01018616 JAC40336.1 GDHF01006374 JAI45940.1 GAKP01018617 JAC40335.1 GDHF01006779 JAI45535.1 AAAB01008960 EAA11668.3 GBXI01006185 JAD08107.1 KQ971326 EEZ97373.2 AGBW02008847 OWR52397.1 OWR52398.1 KQ459601 KPI93701.1 APCN01002262 JN033728 AEW46881.1 GAMC01007113 JAB99442.1 KQ459875 KPJ19634.1 GBXI01000924 JAD13368.1 APGK01033064 APGK01033065 APGK01033066 KB740860 ENN78593.1 BT127127 AEE62089.1 AGBW02011758 OWR46180.1 RSAL01000077 RVE48784.1 KQ459598 KPI94627.1 JXJN01024604 JXJN01024605 JXJN01024606 AB508958 BAI50022.1 EAA44227.3 KQ460779 KPJ12246.1 KZ149936 PZC77130.1 AB508959 BAI50023.1 KPI94624.1 AB508960 BAI50024.1 GEZM01091306 JAV56934.1 KPJ12247.1 AB264710 BAG30785.1 NWSH01000819 PCG74065.1 NEVH01018372 PNF23684.1 EAL40075.2 AB508955 BAI50019.1 ODYU01001971 SOQ38904.1 KPJ12242.1 JN033714 AEW46867.1 KPJ12243.1 GU591172 ADD92156.1 KPJ12244.1 DS232013 EDS31873.1 AB508954 AB508956 BAI50018.1 BAI50020.1 KU170546 APM84101.1 KT992464 AOY34571.1 KC533771 AGP76179.1 ADMH02000892 ETN64737.1 KQ461150 KPJ08731.1 NWSH01004297 PCG65255.1 KC891004 AGS32242.1 AXCM01009805 AB508957 BAI50021.1 KQ459597 KPI94941.1 GGFL01002037 MBW66215.1 GEDC01015759 JAS21539.1 CH477568 EAT38910.1 ETN64734.1
Proteomes
UP000005204
UP000283053
UP000218220
UP000053268
UP000192223
UP000007062
+ More
UP000007266 UP000007151 UP000075903 UP000075840 UP000053240 UP000019118 UP000076407 UP000091820 UP000075880 UP000075882 UP000092460 UP000092443 UP000078200 UP000092445 UP000075901 UP000075920 UP000076408 UP000075902 UP000235965 UP000002320 UP000000673 UP000075883 UP000008820
UP000007266 UP000007151 UP000075903 UP000075840 UP000053240 UP000019118 UP000076407 UP000091820 UP000075880 UP000075882 UP000092460 UP000092443 UP000078200 UP000092445 UP000075901 UP000075920 UP000076408 UP000075902 UP000235965 UP000002320 UP000000673 UP000075883 UP000008820
Interpro
SUPFAM
SSF51445
SSF51445
ProteinModelPortal
H9JX80
H9JX83
H9JXC4
A0A3S2N597
A0A2A4J905
A0A2A4IWH6
+ More
A0A2W1BMB0 A0A194QH27 A0A2W1BC39 H9JX79 A0A2H1W6N8 A0A3G1T1E9 A0A2H1WLJ0 A0A2H1VC49 A0A194QFC2 A0A194QFH9 A0A2A4IZ96 A0A2W1BEU6 H9JXC5 A0A2W1BJB1 H9JXC7 A0A0T6B9C6 A0A0K8UIW8 A0A034VAL4 A0A0K8W4X1 A0A034VEY7 A0A0K8W3Q7 A0A1W4XQQ3 Q7Q5I1 A0A0A1XAG3 D6X3T6 A0A212FF93 A0A1Y9IS68 A0A212FF90 A0A194PJU3 A0A1Y9GLV6 G9F9I3 W8BL15 A0A194RQF8 A0A0A1XS57 N6TKX9 J3JV61 A0A212EXF5 A0A3S2PE71 A0A194PMT2 A0A1Y9J0B2 A0A1A9W4X6 A0A182JGM2 A0A182LL18 A0A1B0C2P0 D0VYR9 A0A1A9XR01 Q7PI98 A0A194R8P7 A0A2W1BWF3 D0VYS0 A0A1A9UP13 A0A194PPG4 A0A1Y9GL87 A0A1A9ZLX0 A0A182TBP9 A0A1Y9IW73 A0A1Y9J0J6 D0VYS1 A0A182YP84 A0A1Y1K6D9 A0A194R3H6 A0A182TWE4 B2DBM6 A0A2A4JPS5 A0A1Y9IS75 A0A2J7Q530 A0A182U0B4 Q5TQY8 A0A1Y9GKT0 A0A1Y9IRY8 D0VYR6 A0A2H1VF65 A0A194R3H3 G9F9G9 A0A194R360 D5KQK3 A0A194R3Z4 B0WNN7 D0VYR5 A0A1S5SJI5 A0A219WGQ6 X2CKZ4 W5JK33 A0A194QYR8 A0A2A4IZK8 S5SP57 A0A182LWK9 D0VYR8 A0A194PNC9 A0A2M4CLR8 A0A1B6D777 Q16WF2 A0A182YP82 W5JPP3
A0A2W1BMB0 A0A194QH27 A0A2W1BC39 H9JX79 A0A2H1W6N8 A0A3G1T1E9 A0A2H1WLJ0 A0A2H1VC49 A0A194QFC2 A0A194QFH9 A0A2A4IZ96 A0A2W1BEU6 H9JXC5 A0A2W1BJB1 H9JXC7 A0A0T6B9C6 A0A0K8UIW8 A0A034VAL4 A0A0K8W4X1 A0A034VEY7 A0A0K8W3Q7 A0A1W4XQQ3 Q7Q5I1 A0A0A1XAG3 D6X3T6 A0A212FF93 A0A1Y9IS68 A0A212FF90 A0A194PJU3 A0A1Y9GLV6 G9F9I3 W8BL15 A0A194RQF8 A0A0A1XS57 N6TKX9 J3JV61 A0A212EXF5 A0A3S2PE71 A0A194PMT2 A0A1Y9J0B2 A0A1A9W4X6 A0A182JGM2 A0A182LL18 A0A1B0C2P0 D0VYR9 A0A1A9XR01 Q7PI98 A0A194R8P7 A0A2W1BWF3 D0VYS0 A0A1A9UP13 A0A194PPG4 A0A1Y9GL87 A0A1A9ZLX0 A0A182TBP9 A0A1Y9IW73 A0A1Y9J0J6 D0VYS1 A0A182YP84 A0A1Y1K6D9 A0A194R3H6 A0A182TWE4 B2DBM6 A0A2A4JPS5 A0A1Y9IS75 A0A2J7Q530 A0A182U0B4 Q5TQY8 A0A1Y9GKT0 A0A1Y9IRY8 D0VYR6 A0A2H1VF65 A0A194R3H3 G9F9G9 A0A194R360 D5KQK3 A0A194R3Z4 B0WNN7 D0VYR5 A0A1S5SJI5 A0A219WGQ6 X2CKZ4 W5JK33 A0A194QYR8 A0A2A4IZK8 S5SP57 A0A182LWK9 D0VYR8 A0A194PNC9 A0A2M4CLR8 A0A1B6D777 Q16WF2 A0A182YP82 W5JPP3
PDB
3VII
E-value=5.86802e-89,
Score=836
Ontologies
GO
PANTHER
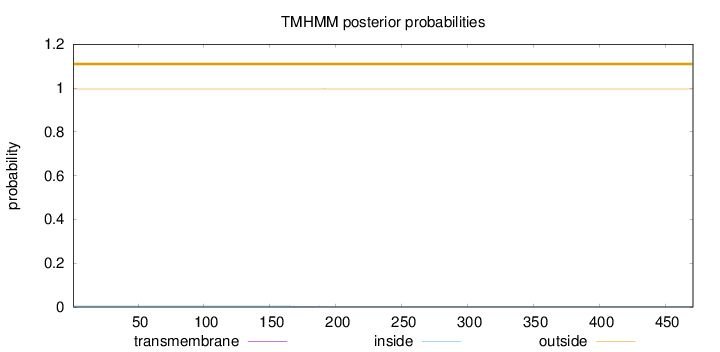
Topology
Length:
471
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01937
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00430
outside
1 - 471
Population Genetic Test Statistics
Pi
230.620742
Theta
200.383791
Tajima's D
2.214927
CLR
0.246242
CSRT
0.914354282285886
Interpretation
Uncertain
