Gene
KWMTBOMO12224 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA014146
Annotation
PREDICTED:_myrosinase_1-like_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 1.536 Nuclear Reliability : 1.323
Sequence
CDS
ATGTTGGTGGTATCTTATGTTCTATGTTTCAGTTCATTCCTTCATCCGCTCTTGGCGAATAAGGAATTGCATTTCCCGAGATGGTTCAAGTTCGGTGCGGCCACGTCGTCGTATCAGATCGAAGGGTCCTGGAATGTTAGTGACAAAACACCAAGTATTTGGGACCATTACACGCATACACAGCCCCATAGGATCAAGGATCACTCCAACGGAGATGTTGCTTGTGACTCTTACAACCAGTACGAGAAGGACATTGAGATGGCCGAGCAGTTGGGACTTGATTTTTATAGGTTTTCTATATCCTGGCCAAGACTGTTGCCTGAGGGATACCGCACTCGCATCAGTTCAGACGGACGGCAATACTACGACAATTTGATCAATGGACTGCTAGACAGGGGGATTGAACCTATGGTCACTTTGTACCATTCAGATATGCCGAAGATATTTCAAGATTTGGGAGGCTGGGCCAATCCTGAAATGGCACAATGGTTTGCCGATTACGCGGACGTAGTATTCTCTCTGTTCGGAGATAGAGTGAAGCTTTGGATCACGTTCAATGAGCCTTTAGTTGCTTGTGATAGCAGCTTACAAACTCTTGGCGTCGTCGAGCCATTCTTGGCTCCATATTTGTGCACGAAGACTGTACTCATGGCTCATGCCAAAGCTTACCATAATTATAAAGATAACTATAAGCCGATGTATCAAGAATCTACTAGATTCAATACCAATAACACTACACGTTCGATGACTTCGGGCTCCAGCAAGCAAGCAAACAAGAAAGCAAGCAAGTTTCCGTGGGGAAGAAAAATTGACACATCTGAAGTCGTCGTGACCTGTAAGATAAGACGTCCGGTAGCCGGAGGTGAGTCTTCCGCGGTCGCGGCCTACCCAGTCCGCGAGAACCAGGCAGATCGCCTCGACGGTACGGAGGCCGGCCGACGGGTCCGCCAAACGACGAGACCACGCCTCCAGCACGGACTGCCGAGATTGGAGCCCCCGCGCTCCAGCTACACTTGCGCTAGGGCGCGCCACCTCGTAGGAGACTGTGAGGTATCCTTGAATGACTCTGACTGCGATCGCACGCTGCCGGCGTCGCAGCGCGGCGACTTTCTCGTGGGTAAGGACGTGGCACCAGACGGGTGCTCCATTCTACTTTGTATTTACGAAGACGGAGCCAGAATACACGGTTACACTTACTGGTCACTGATGGACAACTTCGAATGGCTTTACGGGTACACATCAAAGTTTGGTCTTTACGACGTCGACTTCTCTGACGAGGGCCGGAAGCGCACTCCAAGACTCTCAGCCCAGTACTACTCATACGTTATCAAGAGCAGATCGATCGATTCTAACAAGTGGAAGTCACAGATGGAAGATACAAACGACAACAAAAAACCTATTGATGATGTCGCATAA
Protein
MLVVSYVLCFSSFLHPLLANKELHFPRWFKFGAATSSYQIEGSWNVSDKTPSIWDHYTHTQPHRIKDHSNGDVACDSYNQYEKDIEMAEQLGLDFYRFSISWPRLLPEGYRTRISSDGRQYYDNLINGLLDRGIEPMVTLYHSDMPKIFQDLGGWANPEMAQWFADYADVVFSLFGDRVKLWITFNEPLVACDSSLQTLGVVEPFLAPYLCTKTVLMAHAKAYHNYKDNYKPMYQESTRFNTNNTTRSMTSGSSKQANKKASKFPWGRKIDTSEVVVTCKIRRPVAGGESSAVAAYPVRENQADRLDGTEAGRRVRQTTRPRLQHGLPRLEPPRSSYTCARARHLVGDCEVSLNDSDCDRTLPASQRGDFLVGKDVAPDGCSILLCIYEDGARIHGYTYWSLMDNFEWLYGYTSKFGLYDVDFSDEGRKRTPRLSAQYYSYVIKSRSIDSNKWKSQMEDTNDNKKPIDDVA
Summary
Similarity
Belongs to the glycosyl hydrolase 1 family.
Uniprot
EMBL
Proteomes
Pfam
PF00232 Glyco_hydro_1
Interpro
SUPFAM
SSF51445
SSF51445
ProteinModelPortal
PDB
3VII
E-value=2.79426e-58,
Score=571
Ontologies
PANTHER
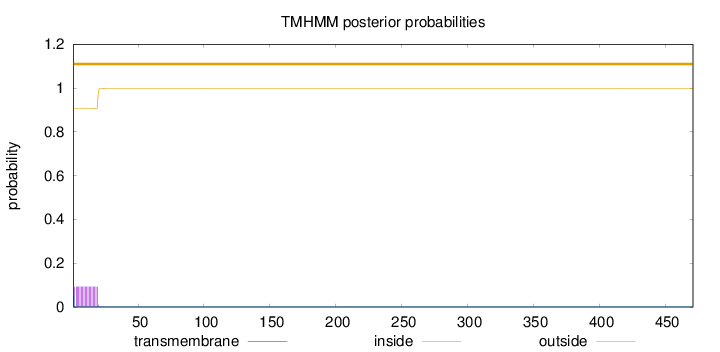
Topology
SignalP
Position: 1 - 19,
Likelihood: 0.679403
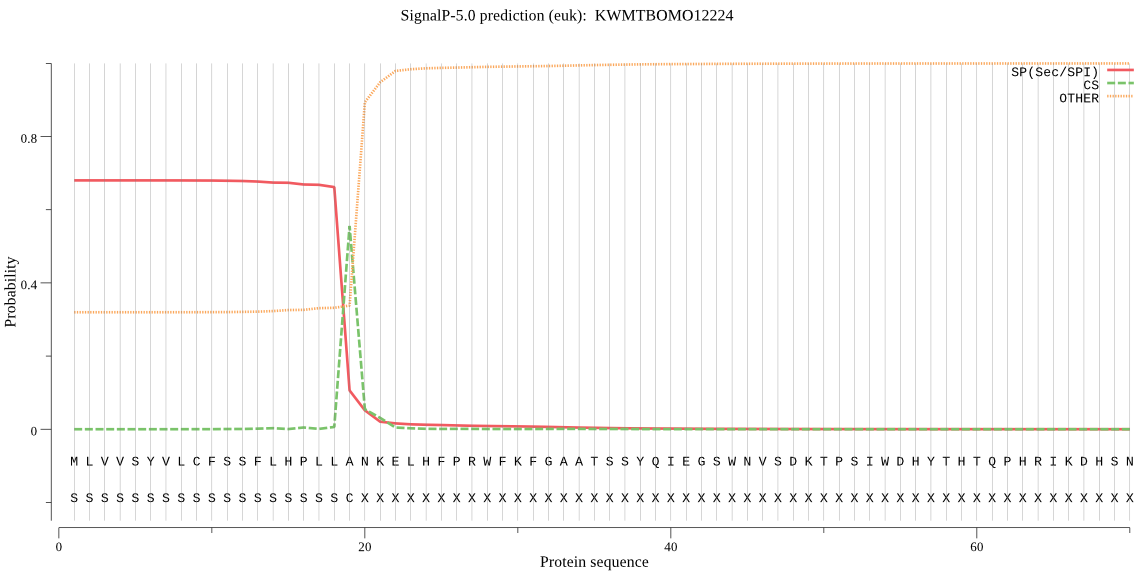
Length:
471
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.69445
Exp number, first 60 AAs:
1.69122
Total prob of N-in:
0.09417
outside
1 - 471
Population Genetic Test Statistics
Pi
255.057371
Theta
179.726193
Tajima's D
0.755019
CLR
0.22138
CSRT
0.591920403979801
Interpretation
Uncertain
