Gene
KWMTBOMO12217
Annotation
PREDICTED:_uncharacterized_protein_LOC101738890_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.428
Sequence
CDS
ATGCTACGTCAAAAAATCATTCAACCCTCTATCTCACCATGGTCAGCACCTATATGGGTAGTACCAAAAAAACAAGACTCCTCCGGTAAACCTAAGTGGCGTATTGTTATCGACTATAGGAAGTTAAATCAAGTAACAATTGGTGATGCCTATCCTATTCCGAACATCACGGATATCCTGGACCAGCTAGGCCACAGTAAATACTTTACCACTCTTGACCTCGCGTCTTCGTTTCATCAAGTCAAGGTGAAAGATACCGAAAAAACAGCATTTAGTGTGCCCTCGGGCCATTACGAGTTCACGCGTATGCCTTTTGGTCTTCGAAATGCTCCTTCCACATTTCAAAGACTCATGAATATAGTACTAAGCGGCCTTCAAGGCACTCATTGCTATACATATTTAGATGATATAGTCGCTTTCAACCACGATCTCTTGTCTCATTGTCAAAACCTCAGAAACATATTTCATAAGCTCCGTGAACACAACCTAAAATTACAACCGGACAAATGTGAGTTCTTTCGTCGAGAAGCACAGTATCTTGGACATATTATTTCAGATAAAGGAGTAATGCCTGACCCCAAGAAGGTTCTCTGTGTTACTAACTTCCCAGTTCCGAAGTCTCCACGTGATATAAAATCATTTCTAGGTTTAGTTGGTTATTATCGTCGCTTCATTGAAAATTTCTCCCATATTACTAAAGCTTTAACTAGCCTTCTAAAAAAGGACGCTACGTTTCAATGGGGTTGTGAACAACAAAAAGCCTTCGACGAACTGAAAAACAAAATAACCTCGGCTCCTATCCTTCAGTACCCTGATTTTACTAAACCCTTCGTACTAACAACAGACGCATCCAATTACGCTGTTTCTGCTATTCTTTCTCAAGGCGAAGTAGGTCAAGATCTCCCAATAGCTTTCGCATCTCGAACCCTGAACAAATCAGAAGGAAATTATTCTACAACCGAAAAAGAATGTTTAGCTATCATTTTCGGCACAAAAGTTTTCAGACCTTATCTGTATGGACACAAGTTTAAAATCGTGACCGATCATAAACCTTTGCAGTGGCTATTTAACTGCAAAGACCCCGGTTCTAAGTTAGTTAGGTGGAGGTTAAAATTAGAAGAATTCGACTACGAGATTGAATACAAAAAGGGGAAGTTAAATTGTAACGCAGATTCCTTATCTCGCAACCTCGTTCATACTCTCGATGAACCAACCGTTTTACAACCACCCCCATACCTTGATGAACAGTCTCGAGTAGATGAGCCCCTTACGTTCGATGACTTGTCTCCGTTACCACCCCCCATAGACTTCAACGCCCCAGGTTCCGTCCCTTCAGAAACCCCTCTTGGACAAGAAATGCCTAGCCACCCTACTCAACCTCCTACAAAAGTCCCTTCTCCATCACTGCCCTCGCAACCGTCACCGTCAACTCCTGAAACCTATGAACAATATCTAAAAGTGTCCCGACAATCGAAAGATCCGTTTAACACAAATATTCAGGAATTTAACGACTCTCTACTTAAAGCTAAATGTAAGCTGATAGCCTACCCTACCTCAATTGACCTTGATGAGTCTGTACCATATTGTTCAGAAATCATAGAAATGTCCACAACTATGCCTGACATACGTGAAGTGGAGAGAGAACTGTACTCATTTTATTCTACCGATTCCAACGACGGTGTCAAGTTCTCTGATGCTATCGACCAAGTACAGTCAGACGAAAAAGCTTTAGCGCATCTTATGCGTGATAATATACATGTGATAAAATCTACAATTTCCACATTTAATAATTCAATGTTAAAATTCAGTGAAAACGAGAAAACGTCTAAATAA
Protein
MLRQKIIQPSISPWSAPIWVVPKKQDSSGKPKWRIVIDYRKLNQVTIGDAYPIPNITDILDQLGHSKYFTTLDLASSFHQVKVKDTEKTAFSVPSGHYEFTRMPFGLRNAPSTFQRLMNIVLSGLQGTHCYTYLDDIVAFNHDLLSHCQNLRNIFHKLREHNLKLQPDKCEFFRREAQYLGHIISDKGVMPDPKKVLCVTNFPVPKSPRDIKSFLGLVGYYRRFIENFSHITKALTSLLKKDATFQWGCEQQKAFDELKNKITSAPILQYPDFTKPFVLTTDASNYAVSAILSQGEVGQDLPIAFASRTLNKSEGNYSTTEKECLAIIFGTKVFRPYLYGHKFKIVTDHKPLQWLFNCKDPGSKLVRWRLKLEEFDYEIEYKKGKLNCNADSLSRNLVHTLDEPTVLQPPPYLDEQSRVDEPLTFDDLSPLPPPIDFNAPGSVPSETPLGQEMPSHPTQPPTKVPSPSLPSQPSPSTPETYEQYLKVSRQSKDPFNTNIQEFNDSLLKAKCKLIAYPTSIDLDESVPYCSEIIEMSTTMPDIREVERELYSFYSTDSNDGVKFSDAIDQVQSDEKALAHLMRDNIHVIKSTISTFNNSMLKFSENEKTSK
Summary
Uniprot
EMBL
Proteomes
Pfam
Interpro
IPR012337
RNaseH-like_sf
+ More
IPR001969 Aspartic_peptidase_AS
IPR001584 Integrase_cat-core
IPR000477 RT_dom
IPR041373 RT_RNaseH
IPR036397 RNaseH_sf
IPR041588 Integrase_H2C2
IPR021109 Peptidase_aspartic_dom_sf
IPR041577 RT_RNaseH_2
IPR002589 Macro_dom
IPR036875 Znf_CCHC_sf
IPR022048 Envelope_fusion-like
IPR003653 Peptidase_C48_C
IPR038765 Papain-like_cys_pep_sf
IPR001969 Aspartic_peptidase_AS
IPR001584 Integrase_cat-core
IPR000477 RT_dom
IPR041373 RT_RNaseH
IPR036397 RNaseH_sf
IPR041588 Integrase_H2C2
IPR021109 Peptidase_aspartic_dom_sf
IPR041577 RT_RNaseH_2
IPR002589 Macro_dom
IPR036875 Znf_CCHC_sf
IPR022048 Envelope_fusion-like
IPR003653 Peptidase_C48_C
IPR038765 Papain-like_cys_pep_sf
Gene 3D
ProteinModelPortal
PDB
4OL8
E-value=3.23788e-68,
Score=658
Ontologies
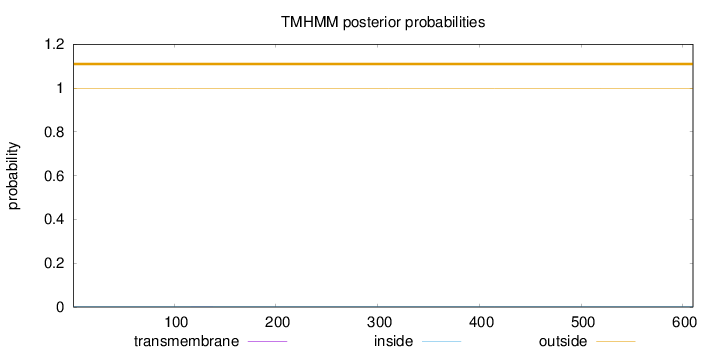
Topology
Length:
610
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00507999999999999
Exp number, first 60 AAs:
0.00035
Total prob of N-in:
0.00107
outside
1 - 610
Population Genetic Test Statistics
Pi
116.479103
Theta
101.524395
Tajima's D
0
CLR
23.068901
CSRT
0.370681465926704
Interpretation
Uncertain
