Gene
KWMTBOMO12208 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA007770
Annotation
PREDICTED:_phosphatidylinositide_phosphatase_SAC1_[Bombyx_mori]
Full name
Phosphatidylinositide phosphatase SAC1
Alternative Name
Suppressor of actin mutations 1-like protein
Location in the cell
PlasmaMembrane Reliability : 2.207
Sequence
CDS
ATGGCAGACCAATTGCTAATTCATGATGATATTAAACTATATACGACATCTGATAAATTTTATTTTGAGCCGACAATCAACCCTACAGAGATTATGGTCATAGACAGAGTCACAGGAGAAGCCAGTGTTAAAGATATAAATTCAGTAAGGATCCCAATACCGGCAAATGCCTACAAACCAGTATGTGGGTTCTTAGGCTCCATAAAACTTATTTCCGGATTGTATTTGGTTGTCGCTAAGTACAGGATACTGGTTGGAAAAATTAATGGGCATGAAATATACCAGCTTGCCGGCACAGACATCATTCCGTATACCAGATCGACTTCACATCTTACTACTCAGCAGATTGCTGACAACAACACGTACGAGCGCATGATCCGGGCTGCACTAGAAACACCGGGAATTTACTTCTCTTACAGCTATGATCTAACTCACACATTGCAGAGACTGCATGCTGTAACACCTGATTTCCATAAGATGTCGCTAGCGAACCGTGCGGACACAAGGTTCCTGTGGAACAGACATCTTCTAAGGGATTACTCCCACGACCAGTTTTCAAGATTCGCTCTGCCCATTATTCAAGGATTTGTGTCGATAAACTATGTGAATATAAACGGGCACGACATGACCTGGGCGCTAGTGTCGCGACGCTGCGCGGATAGAGCCGGCACCAGACTCTTCATGAGGGGCGTCGACGCGCAGGGCAACGTGGCCAATTTTGTAGAAACCGAACAGATAGTTGAGCGCGGCGGCGAGAAGTCTTCGTACGTGCAGACCCGGGGCTCGATCCCGCTGTTCTGGAGCCAGTACCCGGACATTCGGTACAAGCCGCCGATGACGCTGGCCCACGAGGACCACGTCGCGGCCTACACCAAGCACCTCCGCGACCAGATACAACGGTACGGGAACCAGGTGCTGGTTAACTTGATCGACCAGCGTGGGCGCGAGGAGGCGCTGGAGCGTGGGTACCGCGCGGCGGCGGCGGCGGCGGCGCTGCCCGGCGTGCGCTACGAGCCCTTCGACTTCCACGCGGAGTGCCGCGCCATGCGCTACCACCGCCTCGCCGTGCTCACCGACAGGATCGCGCACGAGCAGACGGAATTCGGCTACTTCCTGAGTCGGGGCGGCTCCGTGCTGCTGCGGCAGAGCGGCGTGTTCCGCACCAACTGCGTGGACTGCCTGGACCGGACCAACGTGGTGCAGAGCCTGCTCGCCAAGCTGCATCTGACGGCGCTCTTCAAGCTCCTCAACGTCTCCAGCTCCGACCAACTGGAGACCAACTTCGAACATCTGTTCAATAACGTGTGGGCGGATCACGCGGACGCCATCTCCACGCAGTACTCCGGGACGGGCGCCCTGAAGACGGACTACACGCGCACCGGCAAGCGCACGCACGTCGGTCTGTTGCGCGACGGCATCAACTCCCTCACGCGATACTACAAGAACAACTTCAGCGACGGCTTCAGACAGGACTCAATAGATCTGTTCCTGGGTAAGTACGTGGTCGCTGAGGGCGAGGGTCTGGCCGCCCCGAGCCCTCTGAGGAGAGGACACGACTGGAAATATATCACGCTCCCTTCCGTGCTGCTGGTTGCCATGTCAATGTTCTGCGCCCAAGCGGTGCTGCCGGCGCGCTACTCCAGCGAAGTGCTGCTCTACCTCATGTTCTGGGGGGCGGCTGCTGGAGCGACGCTCTCCTACATCGTCAGGCACGGCGCGGAGTTCGTGGACTGGCCGAGTCTGGACGCCGGGGGCCTGGCGTCGGCGCGCGCCCTCGCCGCCCAGCAGTCTTTATGA
Protein
MADQLLIHDDIKLYTTSDKFYFEPTINPTEIMVIDRVTGEASVKDINSVRIPIPANAYKPVCGFLGSIKLISGLYLVVAKYRILVGKINGHEIYQLAGTDIIPYTRSTSHLTTQQIADNNTYERMIRAALETPGIYFSYSYDLTHTLQRLHAVTPDFHKMSLANRADTRFLWNRHLLRDYSHDQFSRFALPIIQGFVSINYVNINGHDMTWALVSRRCADRAGTRLFMRGVDAQGNVANFVETEQIVERGGEKSSYVQTRGSIPLFWSQYPDIRYKPPMTLAHEDHVAAYTKHLRDQIQRYGNQVLVNLIDQRGREEALERGYRAAAAAAALPGVRYEPFDFHAECRAMRYHRLAVLTDRIAHEQTEFGYFLSRGGSVLLRQSGVFRTNCVDCLDRTNVVQSLLAKLHLTALFKLLNVSSSDQLETNFEHLFNNVWADHADAISTQYSGTGALKTDYTRTGKRTHVGLLRDGINSLTRYYKNNFSDGFRQDSIDLFLGKYVVAEGEGLAAPSPLRRGHDWKYITLPSVLLVAMSMFCAQAVLPARYSSEVLLYLMFWGAAAGATLSYIVRHGAEFVDWPSLDAGGLASARALAAQQSL
Summary
Description
Phosphoinositide phosphatase that hydrolyzes PtdIns(3)P and PtdIns(4)P. Has low activity towards PtdIns(3,5)P2 (By similarity).
Keywords
Complete proteome
Endoplasmic reticulum
Hydrolase
Membrane
Reference proteome
Transmembrane
Transmembrane helix
Feature
chain Phosphatidylinositide phosphatase SAC1
Uniprot
A0A2A4JDW7
A0A3S2NLM3
A0A212F1U3
A0A194QLA8
A0A067RL58
A0A1L8DUN9
+ More
A0A1J1IV69 Q171G2 A0A182G0U6 U5EZ16 A0A2J7QJM1 A0A023EX49 A0A1Q3FCU8 B0WWS3 W8BM43 N6T361 A0A0K8TMQ9 A0A336KBA1 A0A0T6AZ35 D6WSC5 A0A034V7X3 A0A1Y1L0D0 A0A0A1WJC3 A0A0K8UI62 F4WW49 A0A026WZI0 A0A195E655 E9J3E8 A0A195EYY9 E2BMY0 E2AHY8 A0A1B6DBF0 A0A232F776 K7J697 E0VIK7 A0A195CTA0 A0A158NLK1 A0A1B6KAC0 A0A1B6FDT6 E9GCU9 A0A151I126 A0A1B6CC19 A0A1W4WWB0 A0A0L7RGZ3 A0A087ZZ41 A0A2A3EDT3 A0A2P6K9U4 A0A0P6GJG1 A0A087UT65 A0A310SN86 A0A0P4XTN5 A0A0P6CS75 A0A1I8QB92 A0A154PQY3 T1PA60 R7T3T3 A0A0M9A1F3 A0A0L0C227 K1QAG7 A0A0M4EZQ6 W4Y1R6 A0A0K8RAH6 T1KBK4 A0A2P2I606 A0A2S2Q599 B4J2F5 A0A1E1XAC6 B4MXY9 B4HVQ3 A0A3B3CKY1 A0A0J9UBU9 Q29EV1 A0A0R3P8H8 A0A0R3P5P7 T1EGD7 A0A1W4W7V0 A0A1W4VV89 A0A0J9RMY4 A0A1S3DPV1 B4LG85 A0A131XC04 H2LED3 A0A224Z3G2 A0A0P4WGI8 M9MRM4 T1GKC4 Q9W0I6 B4QLY6 A0A3Q0JJ97 A0A3B1K197 A0A2H8TWH0 A0A3B0JRL3 A0A3B0KGA3 A0A2R5LM26 A0A0L8G743 A0A3B0JRK2 A0A3P9J976 G3MLV2 A0A131Z7L7
A0A1J1IV69 Q171G2 A0A182G0U6 U5EZ16 A0A2J7QJM1 A0A023EX49 A0A1Q3FCU8 B0WWS3 W8BM43 N6T361 A0A0K8TMQ9 A0A336KBA1 A0A0T6AZ35 D6WSC5 A0A034V7X3 A0A1Y1L0D0 A0A0A1WJC3 A0A0K8UI62 F4WW49 A0A026WZI0 A0A195E655 E9J3E8 A0A195EYY9 E2BMY0 E2AHY8 A0A1B6DBF0 A0A232F776 K7J697 E0VIK7 A0A195CTA0 A0A158NLK1 A0A1B6KAC0 A0A1B6FDT6 E9GCU9 A0A151I126 A0A1B6CC19 A0A1W4WWB0 A0A0L7RGZ3 A0A087ZZ41 A0A2A3EDT3 A0A2P6K9U4 A0A0P6GJG1 A0A087UT65 A0A310SN86 A0A0P4XTN5 A0A0P6CS75 A0A1I8QB92 A0A154PQY3 T1PA60 R7T3T3 A0A0M9A1F3 A0A0L0C227 K1QAG7 A0A0M4EZQ6 W4Y1R6 A0A0K8RAH6 T1KBK4 A0A2P2I606 A0A2S2Q599 B4J2F5 A0A1E1XAC6 B4MXY9 B4HVQ3 A0A3B3CKY1 A0A0J9UBU9 Q29EV1 A0A0R3P8H8 A0A0R3P5P7 T1EGD7 A0A1W4W7V0 A0A1W4VV89 A0A0J9RMY4 A0A1S3DPV1 B4LG85 A0A131XC04 H2LED3 A0A224Z3G2 A0A0P4WGI8 M9MRM4 T1GKC4 Q9W0I6 B4QLY6 A0A3Q0JJ97 A0A3B1K197 A0A2H8TWH0 A0A3B0JRL3 A0A3B0KGA3 A0A2R5LM26 A0A0L8G743 A0A3B0JRK2 A0A3P9J976 G3MLV2 A0A131Z7L7
EC Number
3.1.3.-
Pubmed
22118469
26354079
24845553
17510324
26483478
24945155
+ More
24495485 23537049 26369729 18362917 19820115 25348373 28004739 25830018 21719571 24508170 30249741 21282665 20798317 28648823 20075255 20566863 21347285 21292972 25315136 23254933 26108605 22992520 17994087 28503490 29451363 22936249 15632085 28049606 17554307 28797301 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 12537569 25329095 22216098 26830274
24495485 23537049 26369729 18362917 19820115 25348373 28004739 25830018 21719571 24508170 30249741 21282665 20798317 28648823 20075255 20566863 21347285 21292972 25315136 23254933 26108605 22992520 17994087 28503490 29451363 22936249 15632085 28049606 17554307 28797301 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 12537569 25329095 22216098 26830274
EMBL
NWSH01001737
PCG70275.1
RSAL01000042
RVE50800.1
AGBW02010795
OWR47702.1
+ More
KQ459053 KPJ04226.1 KK852611 KDR20269.1 GFDF01003935 JAV10149.1 CVRI01000061 CRL04071.1 CH477456 EAT40629.1 JXUM01135349 KQ568258 KXJ69075.1 GANO01001604 JAB58267.1 NEVH01013550 PNF28791.1 GAPW01000649 JAC12949.1 GFDL01009727 JAV25318.1 DS232152 EDS36184.1 GAMC01015816 JAB90739.1 APGK01054085 KB741247 KB632281 ENN71993.1 ERL91330.1 GDAI01001979 JAI15624.1 UFQS01000171 UFQT01000171 SSX00705.1 SSX21085.1 LJIG01022479 KRT80353.1 KQ971352 EFA06386.1 GAKP01019546 JAC39406.1 GEZM01068176 JAV67113.1 GBXI01015677 JAC98614.1 GDHF01026093 JAI26221.1 GL888404 EGI61638.1 KK107063 QOIP01000005 EZA61121.1 RLU22282.1 KQ979579 KYN20680.1 GL768063 EFZ12654.1 KQ981914 KYN33122.1 GL449382 EFN82884.1 GL439602 EFN66978.1 GEDC01014300 JAS22998.1 NNAY01000798 OXU26452.1 DS235200 EEB13213.1 KQ977349 KYN03364.1 ADTU01019612 GEBQ01031586 JAT08391.1 GECZ01021383 JAS48386.1 GL732539 EFX82800.1 KQ976605 KYM79347.1 GEDC01026453 JAS10845.1 KQ414596 KOC70103.1 KZ288269 PBC29887.1 MWRG01019084 PRD23069.1 GDIQ01044762 JAN49975.1 KK121473 KFM80554.1 KQ759820 OAD62735.1 GDIP01240936 JAI82465.1 GDIQ01088091 JAN06646.1 KQ435037 KZC14157.1 KA645622 AFP60251.1 AMQN01003652 KB312379 ELT87336.1 KQ435765 KOX75375.1 JRES01000987 KNC26398.1 JH823241 EKC33652.1 CP012525 ALC44237.1 AAGJ04101869 AAGJ04101870 GADI01005935 JAA67873.1 CAEY01001951 IACF01003849 LAB69457.1 GGMS01003706 MBY72909.1 CH916366 EDV96014.1 GFAC01002965 JAT96223.1 CH963876 EDW76978.1 CH480817 EDW50018.1 CM002912 KMY96775.1 CH379070 EAL29959.2 KRT08517.1 KRT08516.1 AMQM01007866 KB097680 ESN91957.1 KMY96774.1 CH940647 EDW69393.1 GEFH01004102 JAP64479.1 GFPF01009466 MAA20612.1 GDRN01057498 JAI65685.1 AE014296 ADV37453.1 CAQQ02394138 AY047571 CM000363 EDX08772.1 GFXV01006830 MBW18635.1 OUUW01000009 SPP84765.1 SPP84766.1 GGLE01006460 MBY10586.1 KQ423675 KOF72420.1 SPP84764.1 JO842853 AEO34470.1 GEDV01002396 JAP86161.1
KQ459053 KPJ04226.1 KK852611 KDR20269.1 GFDF01003935 JAV10149.1 CVRI01000061 CRL04071.1 CH477456 EAT40629.1 JXUM01135349 KQ568258 KXJ69075.1 GANO01001604 JAB58267.1 NEVH01013550 PNF28791.1 GAPW01000649 JAC12949.1 GFDL01009727 JAV25318.1 DS232152 EDS36184.1 GAMC01015816 JAB90739.1 APGK01054085 KB741247 KB632281 ENN71993.1 ERL91330.1 GDAI01001979 JAI15624.1 UFQS01000171 UFQT01000171 SSX00705.1 SSX21085.1 LJIG01022479 KRT80353.1 KQ971352 EFA06386.1 GAKP01019546 JAC39406.1 GEZM01068176 JAV67113.1 GBXI01015677 JAC98614.1 GDHF01026093 JAI26221.1 GL888404 EGI61638.1 KK107063 QOIP01000005 EZA61121.1 RLU22282.1 KQ979579 KYN20680.1 GL768063 EFZ12654.1 KQ981914 KYN33122.1 GL449382 EFN82884.1 GL439602 EFN66978.1 GEDC01014300 JAS22998.1 NNAY01000798 OXU26452.1 DS235200 EEB13213.1 KQ977349 KYN03364.1 ADTU01019612 GEBQ01031586 JAT08391.1 GECZ01021383 JAS48386.1 GL732539 EFX82800.1 KQ976605 KYM79347.1 GEDC01026453 JAS10845.1 KQ414596 KOC70103.1 KZ288269 PBC29887.1 MWRG01019084 PRD23069.1 GDIQ01044762 JAN49975.1 KK121473 KFM80554.1 KQ759820 OAD62735.1 GDIP01240936 JAI82465.1 GDIQ01088091 JAN06646.1 KQ435037 KZC14157.1 KA645622 AFP60251.1 AMQN01003652 KB312379 ELT87336.1 KQ435765 KOX75375.1 JRES01000987 KNC26398.1 JH823241 EKC33652.1 CP012525 ALC44237.1 AAGJ04101869 AAGJ04101870 GADI01005935 JAA67873.1 CAEY01001951 IACF01003849 LAB69457.1 GGMS01003706 MBY72909.1 CH916366 EDV96014.1 GFAC01002965 JAT96223.1 CH963876 EDW76978.1 CH480817 EDW50018.1 CM002912 KMY96775.1 CH379070 EAL29959.2 KRT08517.1 KRT08516.1 AMQM01007866 KB097680 ESN91957.1 KMY96774.1 CH940647 EDW69393.1 GEFH01004102 JAP64479.1 GFPF01009466 MAA20612.1 GDRN01057498 JAI65685.1 AE014296 ADV37453.1 CAQQ02394138 AY047571 CM000363 EDX08772.1 GFXV01006830 MBW18635.1 OUUW01000009 SPP84765.1 SPP84766.1 GGLE01006460 MBY10586.1 KQ423675 KOF72420.1 SPP84764.1 JO842853 AEO34470.1 GEDV01002396 JAP86161.1
Proteomes
UP000218220
UP000283053
UP000007151
UP000053268
UP000027135
UP000183832
+ More
UP000008820 UP000069940 UP000249989 UP000235965 UP000002320 UP000019118 UP000030742 UP000007266 UP000007755 UP000053097 UP000279307 UP000078492 UP000078541 UP000008237 UP000000311 UP000215335 UP000002358 UP000009046 UP000078542 UP000005205 UP000000305 UP000078540 UP000192223 UP000053825 UP000005203 UP000242457 UP000054359 UP000095300 UP000076502 UP000095301 UP000014760 UP000053105 UP000037069 UP000005408 UP000092553 UP000007110 UP000015104 UP000001070 UP000007798 UP000001292 UP000261560 UP000001819 UP000015101 UP000192221 UP000079169 UP000008792 UP000001038 UP000000803 UP000015102 UP000000304 UP000018467 UP000268350 UP000053454 UP000265200
UP000008820 UP000069940 UP000249989 UP000235965 UP000002320 UP000019118 UP000030742 UP000007266 UP000007755 UP000053097 UP000279307 UP000078492 UP000078541 UP000008237 UP000000311 UP000215335 UP000002358 UP000009046 UP000078542 UP000005205 UP000000305 UP000078540 UP000192223 UP000053825 UP000005203 UP000242457 UP000054359 UP000095300 UP000076502 UP000095301 UP000014760 UP000053105 UP000037069 UP000005408 UP000092553 UP000007110 UP000015104 UP000001070 UP000007798 UP000001292 UP000261560 UP000001819 UP000015101 UP000192221 UP000079169 UP000008792 UP000001038 UP000000803 UP000015102 UP000000304 UP000018467 UP000268350 UP000053454 UP000265200
Pfam
PF02383 Syja_N
Interpro
IPR002013
SAC_dom
ProteinModelPortal
A0A2A4JDW7
A0A3S2NLM3
A0A212F1U3
A0A194QLA8
A0A067RL58
A0A1L8DUN9
+ More
A0A1J1IV69 Q171G2 A0A182G0U6 U5EZ16 A0A2J7QJM1 A0A023EX49 A0A1Q3FCU8 B0WWS3 W8BM43 N6T361 A0A0K8TMQ9 A0A336KBA1 A0A0T6AZ35 D6WSC5 A0A034V7X3 A0A1Y1L0D0 A0A0A1WJC3 A0A0K8UI62 F4WW49 A0A026WZI0 A0A195E655 E9J3E8 A0A195EYY9 E2BMY0 E2AHY8 A0A1B6DBF0 A0A232F776 K7J697 E0VIK7 A0A195CTA0 A0A158NLK1 A0A1B6KAC0 A0A1B6FDT6 E9GCU9 A0A151I126 A0A1B6CC19 A0A1W4WWB0 A0A0L7RGZ3 A0A087ZZ41 A0A2A3EDT3 A0A2P6K9U4 A0A0P6GJG1 A0A087UT65 A0A310SN86 A0A0P4XTN5 A0A0P6CS75 A0A1I8QB92 A0A154PQY3 T1PA60 R7T3T3 A0A0M9A1F3 A0A0L0C227 K1QAG7 A0A0M4EZQ6 W4Y1R6 A0A0K8RAH6 T1KBK4 A0A2P2I606 A0A2S2Q599 B4J2F5 A0A1E1XAC6 B4MXY9 B4HVQ3 A0A3B3CKY1 A0A0J9UBU9 Q29EV1 A0A0R3P8H8 A0A0R3P5P7 T1EGD7 A0A1W4W7V0 A0A1W4VV89 A0A0J9RMY4 A0A1S3DPV1 B4LG85 A0A131XC04 H2LED3 A0A224Z3G2 A0A0P4WGI8 M9MRM4 T1GKC4 Q9W0I6 B4QLY6 A0A3Q0JJ97 A0A3B1K197 A0A2H8TWH0 A0A3B0JRL3 A0A3B0KGA3 A0A2R5LM26 A0A0L8G743 A0A3B0JRK2 A0A3P9J976 G3MLV2 A0A131Z7L7
A0A1J1IV69 Q171G2 A0A182G0U6 U5EZ16 A0A2J7QJM1 A0A023EX49 A0A1Q3FCU8 B0WWS3 W8BM43 N6T361 A0A0K8TMQ9 A0A336KBA1 A0A0T6AZ35 D6WSC5 A0A034V7X3 A0A1Y1L0D0 A0A0A1WJC3 A0A0K8UI62 F4WW49 A0A026WZI0 A0A195E655 E9J3E8 A0A195EYY9 E2BMY0 E2AHY8 A0A1B6DBF0 A0A232F776 K7J697 E0VIK7 A0A195CTA0 A0A158NLK1 A0A1B6KAC0 A0A1B6FDT6 E9GCU9 A0A151I126 A0A1B6CC19 A0A1W4WWB0 A0A0L7RGZ3 A0A087ZZ41 A0A2A3EDT3 A0A2P6K9U4 A0A0P6GJG1 A0A087UT65 A0A310SN86 A0A0P4XTN5 A0A0P6CS75 A0A1I8QB92 A0A154PQY3 T1PA60 R7T3T3 A0A0M9A1F3 A0A0L0C227 K1QAG7 A0A0M4EZQ6 W4Y1R6 A0A0K8RAH6 T1KBK4 A0A2P2I606 A0A2S2Q599 B4J2F5 A0A1E1XAC6 B4MXY9 B4HVQ3 A0A3B3CKY1 A0A0J9UBU9 Q29EV1 A0A0R3P8H8 A0A0R3P5P7 T1EGD7 A0A1W4W7V0 A0A1W4VV89 A0A0J9RMY4 A0A1S3DPV1 B4LG85 A0A131XC04 H2LED3 A0A224Z3G2 A0A0P4WGI8 M9MRM4 T1GKC4 Q9W0I6 B4QLY6 A0A3Q0JJ97 A0A3B1K197 A0A2H8TWH0 A0A3B0JRL3 A0A3B0KGA3 A0A2R5LM26 A0A0L8G743 A0A3B0JRK2 A0A3P9J976 G3MLV2 A0A131Z7L7
PDB
4TU3
E-value=5.31061e-90,
Score=846
Ontologies
PATHWAY
GO
GO:0042578
GO:0016021
GO:0046856
GO:0005783
GO:0043812
GO:0008088
GO:0060074
GO:0046488
GO:0031122
GO:0046329
GO:0030424
GO:0052866
GO:0016199
GO:0043025
GO:0046664
GO:0071683
GO:0005789
GO:0012505
GO:0016791
GO:0035149
GO:0005524
GO:0005515
GO:0003676
GO:0016788
GO:0007165
GO:0005525
GO:0008033
GO:0016787
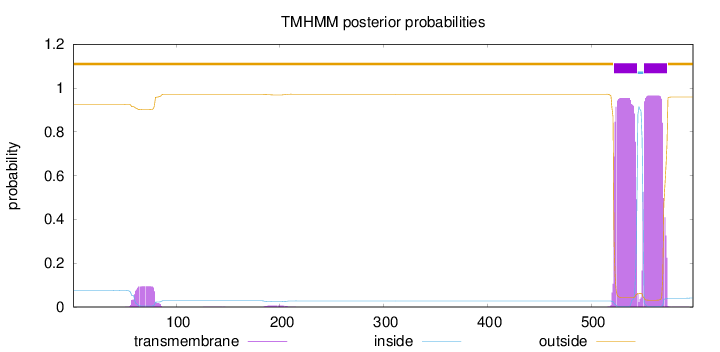
Topology
Subcellular location
Endoplasmic reticulum membrane
Length:
598
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
43.49419
Exp number, first 60 AAs:
0.17079
Total prob of N-in:
0.07477
outside
1 - 521
TMhelix
522 - 544
inside
545 - 550
TMhelix
551 - 573
outside
574 - 598
Population Genetic Test Statistics
Pi
5.664844
Theta
170.206929
Tajima's D
-1.850507
CLR
287.787904
CSRT
0.024298785060747
Interpretation
Possibly Positive selection
