Gene
KWMTBOMO12205 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA014181
Annotation
PREDICTED:_trifunctional_enzyme_subunit_beta?_mitochondrial_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 3.189
Sequence
CDS
ATGGCTTCACAAGTCGCTAAATCATTGATAAAAGCAAACCACGTACGATCTACTGTTAAGTTCGATACTGCCAGACGAGCGCTGAGTGTAGCGCCAGTACTGCAGCAGAAGAAATCTCTCCCAGACCGTACAGGCAAGAATGTGGTGCTTGTCGATGGAGTCCGCACTCCATTCCTTGTTTCATTCACCGATTATGCCAGGATGATGCCCCATGAACTAGCAAGACATGCACTTCTTGGTCTCCTACAAAAGACTGGTATATCGAAGGATGTCATTGACTACATTGTATATGGCACCGTGATCCAAGAGGTGAAAACTTCAAATATAGGGCGCGAGGCAGCCCTGGCGGCTGGGTTCAGCGACAGGACTCCAGCGCACACCGTCACCATGGCTTGCATCTCTTCTAACCAGGCAATCACCACTGGTATCGGTATGATAGCGGCCGGCGCGTATGACGTCATCGTGGCTGGCGGAGTGGAGTTCATGTCGGACGTGCCCATCCGCCACTCGCGCAAGATGCGCTCGCTGCTGCTGCGGATCAACCGCGCCAAGACCCCGGCGCAGAGGCTATCGCTCATTGCCTCCATACGACCGGACTTCTTCGCTCCCGAGCTGCCTGCCGTGGCGGAGTTCTCGTCGGGCGAGACCATGGGCCACAGCGCGGACCGGCTGGCGGCCGCCTTCGGAGCCTCGCGACTGGAGCAGGACCAGTACGCGCTGCGCTCGCACACACTCGCCCACGAGGCCCAGCAGAAGGGATACTTCACGGACCTCATACCGGTCAAAGTGGACGGCAAGGACGGGCTGGTGGACAAGGACAACGGCATCCGCGTGTCCGCCCCCGAGCAGCTCGCCAAGCTGAAGCCGGCCTTCATCAAGCCGCACGGCACCGTCACCGCCGCCAACGCATCCTTCCTGACGGACGGCGCCTCCGCCTGCCTCGTGATGTCCGAGGCGAAGGCCAAGGAGCTCGGCCTCAAACCGAAGGCCTACCTGCGGGACTTCACCTACGTCGCTCAGGACCCCGTGGACCAGCTGCTGCTGGGGCCCGCCTACGGCATTCCCAAGATCTTGGACAAAGTCGGCCTCAAGGTCAACGATATCGACACGTGGGAGATCCATGAGGCCTTTGCGGGACAAATCCTGGCGAACTTGAAGGCTCTCGACTCGGACTGGTTCGGGCAGAAGTATCTGGGACGTCAAGGAAAGGTTGGCAGTCCGGACCTCGAGAAGTGGAACAAGTGGGGCGGCTCGCTCTCTATCGGACATCCATTCGCCGCCACCGGCGTTCGTCTGGCGATGCACACGGCTCACCGGCTGGTGCGGGAGGACGGGCAGTTCGGCGTCATCTCGGCCTGCGCGGCGGGAGGACAGGGAGTCGCCATGCTGCTGGAGAGACATCCTGATGCCACCACCCAATAA
Protein
MASQVAKSLIKANHVRSTVKFDTARRALSVAPVLQQKKSLPDRTGKNVVLVDGVRTPFLVSFTDYARMMPHELARHALLGLLQKTGISKDVIDYIVYGTVIQEVKTSNIGREAALAAGFSDRTPAHTVTMACISSNQAITTGIGMIAAGAYDVIVAGGVEFMSDVPIRHSRKMRSLLLRINRAKTPAQRLSLIASIRPDFFAPELPAVAEFSSGETMGHSADRLAAAFGASRLEQDQYALRSHTLAHEAQQKGYFTDLIPVKVDGKDGLVDKDNGIRVSAPEQLAKLKPAFIKPHGTVTAANASFLTDGASACLVMSEAKAKELGLKPKAYLRDFTYVAQDPVDQLLLGPAYGIPKILDKVGLKVNDIDTWEIHEAFAGQILANLKALDSDWFGQKYLGRQGKVGSPDLEKWNKWGGSLSIGHPFAATGVRLAMHTAHRLVREDGQFGVISACAAGGQGVAMLLERHPDATTQ
Summary
Similarity
Belongs to the thiolase-like superfamily. Thiolase family.
Belongs to the peptidase C1 family.
Belongs to the peptidase C1 family.
Uniprot
H9JXB6
A0A291P1A4
A0A291P143
A0A068FL96
A0A212FF05
A0A3S2NWP4
+ More
D2XML2 S4NU14 A0A2M4A8C4 A0A182JSJ7 A0A182P7H8 A0A2M4BLX6 A0A1S3D5P3 A0A2M3Z1F6 A0A2M4CST8 A0A2M4CSI5 A0A182N3W1 T1DTD7 W5JKK8 A0A182QQ86 A0A182S542 A0A1Q3FHF7 A0A2M4CSD2 A0A182W1X3 Q17IM1 A0A182H0A5 A0A1L8DZE9 A0A182XGS3 A0A023EV11 A0A182VJV1 A0A182HHE0 A0A182FV00 A0A182UJJ1 A0A182LT25 A0A2H1VW16 U5ESQ9 A0A1S4H7M5 B0XFX4 A0A0K8TP31 W8AY77 A0A0P4VKN4 R4FKN5 A0A023F8U1 A0A182J8Z8 A0A336MRZ1 A0A224XPC0 A0A336MAH5 A0A069DUD0 A0A0V0G6N1 A0A0T6BEX1 V5GJ49 A0A1B6CAE5 A0A023ES23 D6WQQ7 A0A1Y1L6R7 A0A1B6EPZ4 B3MCL1 A0A2M3ZGA2 A0A0A1WPJ1 J9K674 A0A1W4V052 B4PA85 B4I8U9 B4QB11 B3NPU1 A0A120MH73 A0A1J1J0K9 A0A2H8TT56 A0A2S2R5H9 B4MRP8 A0A034WF11 Q9W1H8 A0A0K8UCN1 Q292R6 B4GCX3 A0A3B0JIT8 A0A034WAY5 A0A170Y1J7 N6U3E2 O77466 A0A1W4X610 A0A0L0CNA6 B4KMD6 A0A0A9YS83 A0A0M5IY19 A0A146M791 B4LNX7 A0A067RKB2 A0A1I8MDM6 A0A0A1WD76 A0A1I8PJ97 A0A1L8ED69 A0A1A9VU11 A0A2J7QAV1 A0A182SHN5 A0A1B0CR47 A0A1B0A1H2 A0A1B0FL92 A0A1A9XB59 A0A182YL99
D2XML2 S4NU14 A0A2M4A8C4 A0A182JSJ7 A0A182P7H8 A0A2M4BLX6 A0A1S3D5P3 A0A2M3Z1F6 A0A2M4CST8 A0A2M4CSI5 A0A182N3W1 T1DTD7 W5JKK8 A0A182QQ86 A0A182S542 A0A1Q3FHF7 A0A2M4CSD2 A0A182W1X3 Q17IM1 A0A182H0A5 A0A1L8DZE9 A0A182XGS3 A0A023EV11 A0A182VJV1 A0A182HHE0 A0A182FV00 A0A182UJJ1 A0A182LT25 A0A2H1VW16 U5ESQ9 A0A1S4H7M5 B0XFX4 A0A0K8TP31 W8AY77 A0A0P4VKN4 R4FKN5 A0A023F8U1 A0A182J8Z8 A0A336MRZ1 A0A224XPC0 A0A336MAH5 A0A069DUD0 A0A0V0G6N1 A0A0T6BEX1 V5GJ49 A0A1B6CAE5 A0A023ES23 D6WQQ7 A0A1Y1L6R7 A0A1B6EPZ4 B3MCL1 A0A2M3ZGA2 A0A0A1WPJ1 J9K674 A0A1W4V052 B4PA85 B4I8U9 B4QB11 B3NPU1 A0A120MH73 A0A1J1J0K9 A0A2H8TT56 A0A2S2R5H9 B4MRP8 A0A034WF11 Q9W1H8 A0A0K8UCN1 Q292R6 B4GCX3 A0A3B0JIT8 A0A034WAY5 A0A170Y1J7 N6U3E2 O77466 A0A1W4X610 A0A0L0CNA6 B4KMD6 A0A0A9YS83 A0A0M5IY19 A0A146M791 B4LNX7 A0A067RKB2 A0A1I8MDM6 A0A0A1WD76 A0A1I8PJ97 A0A1L8ED69 A0A1A9VU11 A0A2J7QAV1 A0A182SHN5 A0A1B0CR47 A0A1B0A1H2 A0A1B0FL92 A0A1A9XB59 A0A182YL99
Pubmed
19121390
28986331
26385554
22118469
20074338
23622113
+ More
20920257 23761445 17510324 26483478 24945155 12364791 26369729 24495485 27129103 25474469 26334808 18362917 19820115 28004739 17994087 25830018 17550304 22936249 25348373 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 23537049 10191082 26108605 25401762 26823975 24845553 25315136 25244985
20920257 23761445 17510324 26483478 24945155 12364791 26369729 24495485 27129103 25474469 26334808 18362917 19820115 28004739 17994087 25830018 17550304 22936249 25348373 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 23537049 10191082 26108605 25401762 26823975 24845553 25315136 25244985
EMBL
BABH01035733
BABH01035734
MF706183
ATJ44610.1
MF687653
ATJ44579.1
+ More
KJ622108 KJ579211 AID66698.1 AIN34687.1 AGBW02008885 OWR52300.1 RSAL01000042 RVE50797.1 GU205158 ADB57045.1 GAIX01011996 JAA80564.1 GGFK01003694 MBW37015.1 GGFJ01004945 MBW54086.1 GGFM01001570 MBW22321.1 GGFL01004063 MBW68241.1 GGFL01004061 MBW68239.1 GAMD01001019 JAB00572.1 ADMH02000915 ETN64676.1 AXCN02001095 GFDL01008058 JAV26987.1 GGFL01004062 MBW68240.1 CH477238 EAT46539.1 JXUM01022530 KQ560634 KXJ81503.1 GFDF01002429 JAV11655.1 GAPW01001279 JAC12319.1 APCN01002407 AXCM01000489 ODYU01004767 SOQ44998.1 GANO01002246 JAB57625.1 AAAB01008986 DS232970 EDS27093.1 GDAI01001948 JAI15655.1 GAMC01020756 GAMC01020754 GAMC01020751 GAMC01020749 JAB85806.1 GDKW01001685 JAI54910.1 ACPB03011673 GAHY01001768 JAA75742.1 GBBI01000861 JAC17851.1 UFQS01001990 UFQT01001990 SSX12889.1 SSX32331.1 GFTR01006555 JAW09871.1 UFQT01000630 SSX25899.1 GBGD01001329 JAC87560.1 GECL01003191 JAP02933.1 LJIG01001049 KRT85891.1 GALX01004372 JAB64094.1 GEDC01026904 JAS10394.1 GAPW01001570 JAC12028.1 KQ971354 EFA06041.1 GEZM01063157 GEZM01063156 JAV69264.1 GECZ01029771 JAS39998.1 CH902619 EDV36245.1 GGFM01006833 MBW27584.1 GBXI01013691 JAD00601.1 ABLF02034738 CM000158 EDW92411.1 CH480824 EDW57024.1 CM000362 CM002911 EDX08448.1 KMY96142.1 CH954179 EDV56882.1 KT768119 AMH84313.1 CVRI01000064 CRL05332.1 GFXV01004967 MBW16772.1 GGMS01015409 MBY84612.1 CH963850 EDW74787.1 GAKP01006257 JAC52695.1 AE013599 AY069124 AAF47083.1 AAL39269.1 GDHF01027920 JAI24394.1 CM000071 EAL24795.1 CH479181 EDW31511.1 OUUW01000001 SPP73156.1 GAKP01006256 JAC52696.1 GEMB01003799 JAR99454.1 APGK01041389 APGK01041390 KB740993 KB632256 ENN76085.1 ERL90698.1 AB010261 AB010262 BAA32690.1 JRES01000153 KNC33771.1 CH933808 EDW09824.1 GBHO01007707 GBRD01012983 GBRD01011121 GBRD01011119 JAG35897.1 JAG52843.1 CP012524 ALC42611.1 GDHC01002988 JAQ15641.1 CH940648 EDW61146.2 KK852423 KDR24252.1 GBXI01017348 JAC96943.1 GFDG01002276 JAV16523.1 NEVH01016325 PNF25699.1 AJWK01024391 CCAG010020375
KJ622108 KJ579211 AID66698.1 AIN34687.1 AGBW02008885 OWR52300.1 RSAL01000042 RVE50797.1 GU205158 ADB57045.1 GAIX01011996 JAA80564.1 GGFK01003694 MBW37015.1 GGFJ01004945 MBW54086.1 GGFM01001570 MBW22321.1 GGFL01004063 MBW68241.1 GGFL01004061 MBW68239.1 GAMD01001019 JAB00572.1 ADMH02000915 ETN64676.1 AXCN02001095 GFDL01008058 JAV26987.1 GGFL01004062 MBW68240.1 CH477238 EAT46539.1 JXUM01022530 KQ560634 KXJ81503.1 GFDF01002429 JAV11655.1 GAPW01001279 JAC12319.1 APCN01002407 AXCM01000489 ODYU01004767 SOQ44998.1 GANO01002246 JAB57625.1 AAAB01008986 DS232970 EDS27093.1 GDAI01001948 JAI15655.1 GAMC01020756 GAMC01020754 GAMC01020751 GAMC01020749 JAB85806.1 GDKW01001685 JAI54910.1 ACPB03011673 GAHY01001768 JAA75742.1 GBBI01000861 JAC17851.1 UFQS01001990 UFQT01001990 SSX12889.1 SSX32331.1 GFTR01006555 JAW09871.1 UFQT01000630 SSX25899.1 GBGD01001329 JAC87560.1 GECL01003191 JAP02933.1 LJIG01001049 KRT85891.1 GALX01004372 JAB64094.1 GEDC01026904 JAS10394.1 GAPW01001570 JAC12028.1 KQ971354 EFA06041.1 GEZM01063157 GEZM01063156 JAV69264.1 GECZ01029771 JAS39998.1 CH902619 EDV36245.1 GGFM01006833 MBW27584.1 GBXI01013691 JAD00601.1 ABLF02034738 CM000158 EDW92411.1 CH480824 EDW57024.1 CM000362 CM002911 EDX08448.1 KMY96142.1 CH954179 EDV56882.1 KT768119 AMH84313.1 CVRI01000064 CRL05332.1 GFXV01004967 MBW16772.1 GGMS01015409 MBY84612.1 CH963850 EDW74787.1 GAKP01006257 JAC52695.1 AE013599 AY069124 AAF47083.1 AAL39269.1 GDHF01027920 JAI24394.1 CM000071 EAL24795.1 CH479181 EDW31511.1 OUUW01000001 SPP73156.1 GAKP01006256 JAC52696.1 GEMB01003799 JAR99454.1 APGK01041389 APGK01041390 KB740993 KB632256 ENN76085.1 ERL90698.1 AB010261 AB010262 BAA32690.1 JRES01000153 KNC33771.1 CH933808 EDW09824.1 GBHO01007707 GBRD01012983 GBRD01011121 GBRD01011119 JAG35897.1 JAG52843.1 CP012524 ALC42611.1 GDHC01002988 JAQ15641.1 CH940648 EDW61146.2 KK852423 KDR24252.1 GBXI01017348 JAC96943.1 GFDG01002276 JAV16523.1 NEVH01016325 PNF25699.1 AJWK01024391 CCAG010020375
Proteomes
UP000005204
UP000007151
UP000283053
UP000075881
UP000075885
UP000079169
+ More
UP000075884 UP000000673 UP000075886 UP000075900 UP000075920 UP000008820 UP000069940 UP000249989 UP000076407 UP000075903 UP000075840 UP000069272 UP000075902 UP000075883 UP000002320 UP000015103 UP000075880 UP000007266 UP000007801 UP000007819 UP000192221 UP000002282 UP000001292 UP000000304 UP000008711 UP000183832 UP000007798 UP000000803 UP000001819 UP000008744 UP000268350 UP000019118 UP000030742 UP000192223 UP000037069 UP000009192 UP000092553 UP000008792 UP000027135 UP000095301 UP000095300 UP000078200 UP000235965 UP000075901 UP000092461 UP000092445 UP000092444 UP000092443 UP000076408
UP000075884 UP000000673 UP000075886 UP000075900 UP000075920 UP000008820 UP000069940 UP000249989 UP000076407 UP000075903 UP000075840 UP000069272 UP000075902 UP000075883 UP000002320 UP000015103 UP000075880 UP000007266 UP000007801 UP000007819 UP000192221 UP000002282 UP000001292 UP000000304 UP000008711 UP000183832 UP000007798 UP000000803 UP000001819 UP000008744 UP000268350 UP000019118 UP000030742 UP000192223 UP000037069 UP000009192 UP000092553 UP000008792 UP000027135 UP000095301 UP000095300 UP000078200 UP000235965 UP000075901 UP000092461 UP000092445 UP000092444 UP000092443 UP000076408
Pfam
Interpro
IPR002155
Thiolase
+ More
IPR020613 Thiolase_CS
IPR020610 Thiolase_AS
IPR020616 Thiolase_N
IPR020617 Thiolase_C
IPR016039 Thiolase-like
IPR020615 Thiolase_acyl_enz_int_AS
IPR025661 Pept_asp_AS
IPR000668 Peptidase_C1A_C
IPR000169 Pept_cys_AS
IPR001614 Myelin_PLP
IPR013201 Prot_inhib_I29
IPR025660 Pept_his_AS
IPR038765 Papain-like_cys_pep_sf
IPR039417 Peptidase_C1A_papain-like
IPR027917 SMIM20
IPR006689 Small_GTPase_ARF/SAR
IPR027417 P-loop_NTPase
IPR005225 Small_GTP-bd_dom
IPR020613 Thiolase_CS
IPR020610 Thiolase_AS
IPR020616 Thiolase_N
IPR020617 Thiolase_C
IPR016039 Thiolase-like
IPR020615 Thiolase_acyl_enz_int_AS
IPR025661 Pept_asp_AS
IPR000668 Peptidase_C1A_C
IPR000169 Pept_cys_AS
IPR001614 Myelin_PLP
IPR013201 Prot_inhib_I29
IPR025660 Pept_his_AS
IPR038765 Papain-like_cys_pep_sf
IPR039417 Peptidase_C1A_papain-like
IPR027917 SMIM20
IPR006689 Small_GTPase_ARF/SAR
IPR027417 P-loop_NTPase
IPR005225 Small_GTP-bd_dom
Gene 3D
ProteinModelPortal
H9JXB6
A0A291P1A4
A0A291P143
A0A068FL96
A0A212FF05
A0A3S2NWP4
+ More
D2XML2 S4NU14 A0A2M4A8C4 A0A182JSJ7 A0A182P7H8 A0A2M4BLX6 A0A1S3D5P3 A0A2M3Z1F6 A0A2M4CST8 A0A2M4CSI5 A0A182N3W1 T1DTD7 W5JKK8 A0A182QQ86 A0A182S542 A0A1Q3FHF7 A0A2M4CSD2 A0A182W1X3 Q17IM1 A0A182H0A5 A0A1L8DZE9 A0A182XGS3 A0A023EV11 A0A182VJV1 A0A182HHE0 A0A182FV00 A0A182UJJ1 A0A182LT25 A0A2H1VW16 U5ESQ9 A0A1S4H7M5 B0XFX4 A0A0K8TP31 W8AY77 A0A0P4VKN4 R4FKN5 A0A023F8U1 A0A182J8Z8 A0A336MRZ1 A0A224XPC0 A0A336MAH5 A0A069DUD0 A0A0V0G6N1 A0A0T6BEX1 V5GJ49 A0A1B6CAE5 A0A023ES23 D6WQQ7 A0A1Y1L6R7 A0A1B6EPZ4 B3MCL1 A0A2M3ZGA2 A0A0A1WPJ1 J9K674 A0A1W4V052 B4PA85 B4I8U9 B4QB11 B3NPU1 A0A120MH73 A0A1J1J0K9 A0A2H8TT56 A0A2S2R5H9 B4MRP8 A0A034WF11 Q9W1H8 A0A0K8UCN1 Q292R6 B4GCX3 A0A3B0JIT8 A0A034WAY5 A0A170Y1J7 N6U3E2 O77466 A0A1W4X610 A0A0L0CNA6 B4KMD6 A0A0A9YS83 A0A0M5IY19 A0A146M791 B4LNX7 A0A067RKB2 A0A1I8MDM6 A0A0A1WD76 A0A1I8PJ97 A0A1L8ED69 A0A1A9VU11 A0A2J7QAV1 A0A182SHN5 A0A1B0CR47 A0A1B0A1H2 A0A1B0FL92 A0A1A9XB59 A0A182YL99
D2XML2 S4NU14 A0A2M4A8C4 A0A182JSJ7 A0A182P7H8 A0A2M4BLX6 A0A1S3D5P3 A0A2M3Z1F6 A0A2M4CST8 A0A2M4CSI5 A0A182N3W1 T1DTD7 W5JKK8 A0A182QQ86 A0A182S542 A0A1Q3FHF7 A0A2M4CSD2 A0A182W1X3 Q17IM1 A0A182H0A5 A0A1L8DZE9 A0A182XGS3 A0A023EV11 A0A182VJV1 A0A182HHE0 A0A182FV00 A0A182UJJ1 A0A182LT25 A0A2H1VW16 U5ESQ9 A0A1S4H7M5 B0XFX4 A0A0K8TP31 W8AY77 A0A0P4VKN4 R4FKN5 A0A023F8U1 A0A182J8Z8 A0A336MRZ1 A0A224XPC0 A0A336MAH5 A0A069DUD0 A0A0V0G6N1 A0A0T6BEX1 V5GJ49 A0A1B6CAE5 A0A023ES23 D6WQQ7 A0A1Y1L6R7 A0A1B6EPZ4 B3MCL1 A0A2M3ZGA2 A0A0A1WPJ1 J9K674 A0A1W4V052 B4PA85 B4I8U9 B4QB11 B3NPU1 A0A120MH73 A0A1J1J0K9 A0A2H8TT56 A0A2S2R5H9 B4MRP8 A0A034WF11 Q9W1H8 A0A0K8UCN1 Q292R6 B4GCX3 A0A3B0JIT8 A0A034WAY5 A0A170Y1J7 N6U3E2 O77466 A0A1W4X610 A0A0L0CNA6 B4KMD6 A0A0A9YS83 A0A0M5IY19 A0A146M791 B4LNX7 A0A067RKB2 A0A1I8MDM6 A0A0A1WD76 A0A1I8PJ97 A0A1L8ED69 A0A1A9VU11 A0A2J7QAV1 A0A182SHN5 A0A1B0CR47 A0A1B0A1H2 A0A1B0FL92 A0A1A9XB59 A0A182YL99
PDB
5ZRV
E-value=3.96181e-168,
Score=1519
Ontologies
PATHWAY
00062
Fatty acid elongation - Bombyx mori (domestic silkworm)
00071 Fatty acid degradation - Bombyx mori (domestic silkworm)
00280 Valine, leucine and isoleucine degradation - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
01212 Fatty acid metabolism - Bombyx mori (domestic silkworm)
00071 Fatty acid degradation - Bombyx mori (domestic silkworm)
00280 Valine, leucine and isoleucine degradation - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
01212 Fatty acid metabolism - Bombyx mori (domestic silkworm)
GO
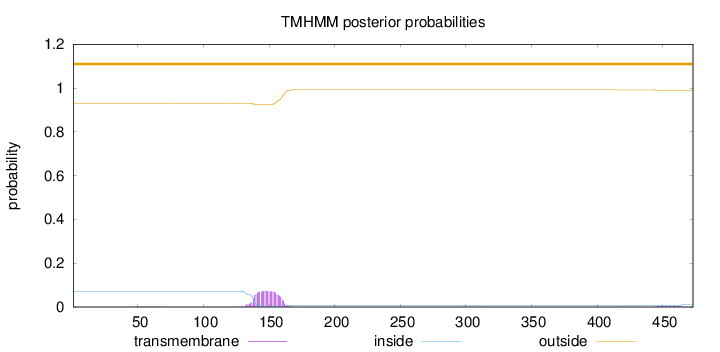
Topology
Length:
473
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.71951
Exp number, first 60 AAs:
0.00655
Total prob of N-in:
0.07158
outside
1 - 473
Population Genetic Test Statistics
Pi
246.777722
Theta
173.910643
Tajima's D
1.929115
CLR
0.100579
CSRT
0.868756562171891
Interpretation
Uncertain
