Gene
KWMTBOMO12204
Pre Gene Modal
BGIBMGA014161
Annotation
vitellogenin_receptor_precursor_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 4.504
Sequence
CDS
ATGAAGGTAGTTTTGTTAGCAATAGTTCTATGTACAACCTCGTGCGCGGGGCAGTTCGTTGACGAAATGCAAGTCTACGAGAAGGAGTGCCTGGGCGAGGATGTGTTTCCGTGCATGTCCGGGGGATGCATACAGCAGTCCCAGTACTGCGACGGAAAGGTGGACTGCGACGATGGAACCGACGAGAACTATTGCCTTGATCACAAGCCAGACGCTCAGTTCTGTAACGAGACCCACCAGTTCATGTGTCGGGACAGCAAGAAGTGCATCCCGAACCATTGGATCTGTAATAACGACATCGATTGCGACGACGGAAGTGATGAGCTAAATTGCACTTTGGTTCCTGTCGCTACTGGTAAATGCAAAGGTTTCCTGTGCGGCGATGGAAAATGTATCTCCAGTCTTTGGTTATGTGATGGAAGCTACGACTGCAAGGATAAGAGCGACGAGAATTCACCGGAGAACTGCCGTCACAGCCTCCTGTCCCACTCGATGCTAAGCGGATCGGATTGCCCGGAGTGGCTAGGAGGGAGGCGCCAATACAAATGCACGGACTCCTCGTTTTGCCTCCCGAGTGAAATGATGTGTGATGGGATGCAGGACTGCAAGGACGGCAGTGACGAGAGACCCTTCTGCGCCAACTGGCACACGATGTGCGCGAACCACACGTGCCTCGGTGACAAGGCCTCGTGTGTGCCGGACCGCGCCGGGCCCACGTGCGAGTGTCTCAACCACCTCAACCTGCGTCGGTACAATACCTCGACCGGGGCCTGCGACGACATCGACGAGTGCGCGCTGGCCCGCCCTCAGTGCTCCCACTACTGCGTCAACGCGGACGGCCATTTCACTTGTGAATGCGCCGACGGCTACTTCAAGGACGAACTTAAGTACTTGTGCTACGCTACCGGTCCCGAACCCCTGTTGTTCTACAGTACACGAAACGAAATTAAATATCTGAAAGTGAAGTCGAAGGAAGTGGTCACACTGGCGACTGGAATAAAAAAGGCTCACGGGGTCACATCGAACGGAATATACGTTTACTGGGTGGAAACAGCTGAAGGTCATCAAGCCATCGTCAAAGCTCACATAGACGACGTAGAAAACACTCGACAGGTTTGTCTTATGTCTTTTTAG
Protein
MKVVLLAIVLCTTSCAGQFVDEMQVYEKECLGEDVFPCMSGGCIQQSQYCDGKVDCDDGTDENYCLDHKPDAQFCNETHQFMCRDSKKCIPNHWICNNDIDCDDGSDELNCTLVPVATGKCKGFLCGDGKCISSLWLCDGSYDCKDKSDENSPENCRHSLLSHSMLSGSDCPEWLGGRRQYKCTDSSFCLPSEMMCDGMQDCKDGSDERPFCANWHTMCANHTCLGDKASCVPDRAGPTCECLNHLNLRRYNTSTGACDDIDECALARPQCSHYCVNADGHFTCECADGYFKDELKYLCYATGPEPLLFYSTRNEIKYLKVKSKEVVTLATGIKKAHGVTSNGIYVYWVETAEGHQAIVKAHIDDVENTRQVCLMSF
Summary
Uniprot
E2FLQ4
H2CLV8
K4MUW4
A0A2A4JFQ2
M1L4G2
A0A385LM65
+ More
A0A2D0VP24 D9ZFI0 A0A194QLA3 A0A2W1BNV3 A0A1Z2RRL4 A0A0N1PFQ7 A0A385HW27 A0A2H1VW67 A0A212FEY0 A0A2S1XV31 A0A1L8E338 T1H951 A0A1B0CK10 A0A0P4ZPP0 A0A0L0C1S0 A0A0P5NI37 A0A0P5JXZ9 A0A0P5PFI2 A0A0P5LY81 A0A3P8NEL8 A0A0P5J3H5 A0A0P5GF13 A0A0P5G933 A0A0N8BJY3 A0A0P6H3Y9 A0A0N8DXE3 A0A3P8NEN0 A0A0P5RWQ5 A0A0P5FXD2 A0A0N8AUY6 A0A0P5IV54 A0A0P5RZH8 A0A0P5PN39 A0A3P9AXL2 A0A0P5DXE5 A0A3P9AXR5 A0A0P5GPI4 A0A0P5L7M2 A0A0P6E3V7 A0A0P5K179 A0A3P8NET0 A0A0P5J1F9 A0A0P5SE30 A0A0P4Z186 A0A0P5I355 A0A0N8CGL0 A0A0P6DFB9 A0A0P5CR20 A0A0P5Z3A9 A0A0P5EGI9 A0A0P5KZS4 A0A0P6E9J7 A0A0P5K6Y3 A0A0P5Q0G8 A0A0N8C0M5 Q2YI44 A0A0P6A4V9 A0A3Q3BHZ1 A0A3B4FMG6 A0A0N8BKC7 A0A1B1UZF6 A0A1B6KIS9 A0A0P5KAT0 A0A023FWP6 A0A3B4BJ67 A0A0P5AT73 A0A0P5LKJ2 A0A0P5FUT7 A0A0P6GYI7 A0A0P6B1N1 A0A0P5LPV1 A0A0P5KGH2 A0A0P4YFW2 A0A0T6B1R3 A0A1B6JXC2 Q1T728 A0A1A9V729 Q45VP9 A0A2J7QQP4 E9H6F1 A0A1B6I2G9 B3S2H0 A0A0P5EP04 A0A369SSL3 A0A3Q2LBF6 A0A1I8MWR0 I3KFZ1 I3KFY9 S7P8B8 I3KFZ0
A0A2D0VP24 D9ZFI0 A0A194QLA3 A0A2W1BNV3 A0A1Z2RRL4 A0A0N1PFQ7 A0A385HW27 A0A2H1VW67 A0A212FEY0 A0A2S1XV31 A0A1L8E338 T1H951 A0A1B0CK10 A0A0P4ZPP0 A0A0L0C1S0 A0A0P5NI37 A0A0P5JXZ9 A0A0P5PFI2 A0A0P5LY81 A0A3P8NEL8 A0A0P5J3H5 A0A0P5GF13 A0A0P5G933 A0A0N8BJY3 A0A0P6H3Y9 A0A0N8DXE3 A0A3P8NEN0 A0A0P5RWQ5 A0A0P5FXD2 A0A0N8AUY6 A0A0P5IV54 A0A0P5RZH8 A0A0P5PN39 A0A3P9AXL2 A0A0P5DXE5 A0A3P9AXR5 A0A0P5GPI4 A0A0P5L7M2 A0A0P6E3V7 A0A0P5K179 A0A3P8NET0 A0A0P5J1F9 A0A0P5SE30 A0A0P4Z186 A0A0P5I355 A0A0N8CGL0 A0A0P6DFB9 A0A0P5CR20 A0A0P5Z3A9 A0A0P5EGI9 A0A0P5KZS4 A0A0P6E9J7 A0A0P5K6Y3 A0A0P5Q0G8 A0A0N8C0M5 Q2YI44 A0A0P6A4V9 A0A3Q3BHZ1 A0A3B4FMG6 A0A0N8BKC7 A0A1B1UZF6 A0A1B6KIS9 A0A0P5KAT0 A0A023FWP6 A0A3B4BJ67 A0A0P5AT73 A0A0P5LKJ2 A0A0P5FUT7 A0A0P6GYI7 A0A0P6B1N1 A0A0P5LPV1 A0A0P5KGH2 A0A0P4YFW2 A0A0T6B1R3 A0A1B6JXC2 Q1T728 A0A1A9V729 Q45VP9 A0A2J7QQP4 E9H6F1 A0A1B6I2G9 B3S2H0 A0A0P5EP04 A0A369SSL3 A0A3Q2LBF6 A0A1I8MWR0 I3KFZ1 I3KFY9 S7P8B8 I3KFZ0
Pubmed
EMBL
HM172611
ADK94452.1
JN003583
AEJ88360.1
JQ809472
AFV32171.1
+ More
NWSH01001747 PCG70260.1 KC181922 AGF33811.2 MH745217 AYA29178.2 KT899978 AOX13593.1 GU983858 ADK94033.1 KQ459053 KPJ04221.1 KZ150016 PZC74967.1 KY963162 ASA46482.1 KQ461113 KPJ08910.1 MG799569 AXY55007.1 ODYU01004767 SOQ44996.1 AGBW02008885 OWR52301.1 MF622539 AWJ95281.1 GFDF01000931 JAV13153.1 ACPB03011459 ACPB03011460 AJWK01015600 GDIP01209607 JAJ13795.1 JRES01000996 KNC26298.1 GDIQ01142056 JAL09670.1 GDIQ01197433 JAK54292.1 GDIQ01129311 JAL22415.1 GDIQ01163447 JAK88278.1 GDIQ01205984 JAK45741.1 GDIQ01241820 JAK09905.1 GDIQ01244480 JAK07245.1 GDIQ01191238 GDIQ01170395 GDIQ01033501 JAK81330.1 GDIQ01037393 JAN57344.1 GDIQ01251522 GDIQ01082524 JAN12213.1 GDIQ01113113 JAL38613.1 GDIQ01250068 JAK01657.1 GDIQ01240371 JAK11354.1 GDIQ01207378 JAK44347.1 GDIQ01099120 JAL52606.1 GDIQ01128015 JAL23711.1 GDIP01149466 JAJ73936.1 GDIQ01238676 JAK13049.1 GDIQ01198837 JAK52888.1 GDIQ01068184 JAN26553.1 GDIQ01191237 JAK60488.1 GDIQ01204480 JAK47245.1 GDIP01141118 JAL62596.1 GDIP01231612 JAI91789.1 GDIQ01225222 JAK26503.1 GDIP01133767 JAL69947.1 GDIQ01087271 JAN07466.1 GDIP01166911 JAJ56491.1 GDIP01049779 JAM53936.1 GDIP01146765 JAJ76637.1 GDIQ01202631 JAK49094.1 GDIQ01066790 JAN27947.1 GDIQ01189892 JAK61833.1 GDIQ01128014 JAL23712.1 GDIQ01126459 JAL25267.1 AM050637 CAJ19121.1 GDIP01034982 JAM68733.1 GDIQ01169243 JAK82482.1 KX428486 ANW09597.1 GEBQ01028624 JAT11353.1 GDIQ01186985 JAK64740.1 GBBL01001034 JAC26286.1 GDIP01194461 JAJ28941.1 GDIQ01168296 JAK83429.1 GDIP01143319 JAJ80083.1 GDIQ01026850 JAN67887.1 GDIP01021304 JAM82411.1 GDIQ01191239 JAK60486.1 GDIQ01191240 JAK60485.1 GDIP01231611 JAI91790.1 LJIG01016164 KRT81424.1 GECU01003850 JAT03857.1 AB255883 BAE93218.1 DQ103506 AAZ31260.3 NEVH01012083 PNF30911.1 GL732597 EFX72644.1 GECU01026651 JAS81055.1 DS985247 EDV23416.1 GDIP01145271 JAJ78131.1 NOWV01000001 RDD47933.1 AERX01019876 AERX01019877 AERX01019878 KE162005 EPQ06388.1
NWSH01001747 PCG70260.1 KC181922 AGF33811.2 MH745217 AYA29178.2 KT899978 AOX13593.1 GU983858 ADK94033.1 KQ459053 KPJ04221.1 KZ150016 PZC74967.1 KY963162 ASA46482.1 KQ461113 KPJ08910.1 MG799569 AXY55007.1 ODYU01004767 SOQ44996.1 AGBW02008885 OWR52301.1 MF622539 AWJ95281.1 GFDF01000931 JAV13153.1 ACPB03011459 ACPB03011460 AJWK01015600 GDIP01209607 JAJ13795.1 JRES01000996 KNC26298.1 GDIQ01142056 JAL09670.1 GDIQ01197433 JAK54292.1 GDIQ01129311 JAL22415.1 GDIQ01163447 JAK88278.1 GDIQ01205984 JAK45741.1 GDIQ01241820 JAK09905.1 GDIQ01244480 JAK07245.1 GDIQ01191238 GDIQ01170395 GDIQ01033501 JAK81330.1 GDIQ01037393 JAN57344.1 GDIQ01251522 GDIQ01082524 JAN12213.1 GDIQ01113113 JAL38613.1 GDIQ01250068 JAK01657.1 GDIQ01240371 JAK11354.1 GDIQ01207378 JAK44347.1 GDIQ01099120 JAL52606.1 GDIQ01128015 JAL23711.1 GDIP01149466 JAJ73936.1 GDIQ01238676 JAK13049.1 GDIQ01198837 JAK52888.1 GDIQ01068184 JAN26553.1 GDIQ01191237 JAK60488.1 GDIQ01204480 JAK47245.1 GDIP01141118 JAL62596.1 GDIP01231612 JAI91789.1 GDIQ01225222 JAK26503.1 GDIP01133767 JAL69947.1 GDIQ01087271 JAN07466.1 GDIP01166911 JAJ56491.1 GDIP01049779 JAM53936.1 GDIP01146765 JAJ76637.1 GDIQ01202631 JAK49094.1 GDIQ01066790 JAN27947.1 GDIQ01189892 JAK61833.1 GDIQ01128014 JAL23712.1 GDIQ01126459 JAL25267.1 AM050637 CAJ19121.1 GDIP01034982 JAM68733.1 GDIQ01169243 JAK82482.1 KX428486 ANW09597.1 GEBQ01028624 JAT11353.1 GDIQ01186985 JAK64740.1 GBBL01001034 JAC26286.1 GDIP01194461 JAJ28941.1 GDIQ01168296 JAK83429.1 GDIP01143319 JAJ80083.1 GDIQ01026850 JAN67887.1 GDIP01021304 JAM82411.1 GDIQ01191239 JAK60486.1 GDIQ01191240 JAK60485.1 GDIP01231611 JAI91790.1 LJIG01016164 KRT81424.1 GECU01003850 JAT03857.1 AB255883 BAE93218.1 DQ103506 AAZ31260.3 NEVH01012083 PNF30911.1 GL732597 EFX72644.1 GECU01026651 JAS81055.1 DS985247 EDV23416.1 GDIP01145271 JAJ78131.1 NOWV01000001 RDD47933.1 AERX01019876 AERX01019877 AERX01019878 KE162005 EPQ06388.1
Proteomes
Interpro
IPR001881
EGF-like_Ca-bd_dom
+ More
IPR000033 LDLR_classB_rpt
IPR036055 LDL_receptor-like_sf
IPR002172 LDrepeatLR_classA_rpt
IPR018097 EGF_Ca-bd_CS
IPR013032 EGF-like_CS
IPR040093 LRAD1
IPR011042 6-blade_b-propeller_TolB-like
IPR000742 EGF-like_dom
IPR000152 EGF-type_Asp/Asn_hydroxyl_site
IPR023415 LDLR_class-A_CS
IPR009030 Growth_fac_rcpt_cys_sf
IPR026823 cEGF
IPR001190 SRCR
IPR032931 VLDLR
IPR000033 LDLR_classB_rpt
IPR036055 LDL_receptor-like_sf
IPR002172 LDrepeatLR_classA_rpt
IPR018097 EGF_Ca-bd_CS
IPR013032 EGF-like_CS
IPR040093 LRAD1
IPR011042 6-blade_b-propeller_TolB-like
IPR000742 EGF-like_dom
IPR000152 EGF-type_Asp/Asn_hydroxyl_site
IPR023415 LDLR_class-A_CS
IPR009030 Growth_fac_rcpt_cys_sf
IPR026823 cEGF
IPR001190 SRCR
IPR032931 VLDLR
Gene 3D
CDD
ProteinModelPortal
E2FLQ4
H2CLV8
K4MUW4
A0A2A4JFQ2
M1L4G2
A0A385LM65
+ More
A0A2D0VP24 D9ZFI0 A0A194QLA3 A0A2W1BNV3 A0A1Z2RRL4 A0A0N1PFQ7 A0A385HW27 A0A2H1VW67 A0A212FEY0 A0A2S1XV31 A0A1L8E338 T1H951 A0A1B0CK10 A0A0P4ZPP0 A0A0L0C1S0 A0A0P5NI37 A0A0P5JXZ9 A0A0P5PFI2 A0A0P5LY81 A0A3P8NEL8 A0A0P5J3H5 A0A0P5GF13 A0A0P5G933 A0A0N8BJY3 A0A0P6H3Y9 A0A0N8DXE3 A0A3P8NEN0 A0A0P5RWQ5 A0A0P5FXD2 A0A0N8AUY6 A0A0P5IV54 A0A0P5RZH8 A0A0P5PN39 A0A3P9AXL2 A0A0P5DXE5 A0A3P9AXR5 A0A0P5GPI4 A0A0P5L7M2 A0A0P6E3V7 A0A0P5K179 A0A3P8NET0 A0A0P5J1F9 A0A0P5SE30 A0A0P4Z186 A0A0P5I355 A0A0N8CGL0 A0A0P6DFB9 A0A0P5CR20 A0A0P5Z3A9 A0A0P5EGI9 A0A0P5KZS4 A0A0P6E9J7 A0A0P5K6Y3 A0A0P5Q0G8 A0A0N8C0M5 Q2YI44 A0A0P6A4V9 A0A3Q3BHZ1 A0A3B4FMG6 A0A0N8BKC7 A0A1B1UZF6 A0A1B6KIS9 A0A0P5KAT0 A0A023FWP6 A0A3B4BJ67 A0A0P5AT73 A0A0P5LKJ2 A0A0P5FUT7 A0A0P6GYI7 A0A0P6B1N1 A0A0P5LPV1 A0A0P5KGH2 A0A0P4YFW2 A0A0T6B1R3 A0A1B6JXC2 Q1T728 A0A1A9V729 Q45VP9 A0A2J7QQP4 E9H6F1 A0A1B6I2G9 B3S2H0 A0A0P5EP04 A0A369SSL3 A0A3Q2LBF6 A0A1I8MWR0 I3KFZ1 I3KFY9 S7P8B8 I3KFZ0
A0A2D0VP24 D9ZFI0 A0A194QLA3 A0A2W1BNV3 A0A1Z2RRL4 A0A0N1PFQ7 A0A385HW27 A0A2H1VW67 A0A212FEY0 A0A2S1XV31 A0A1L8E338 T1H951 A0A1B0CK10 A0A0P4ZPP0 A0A0L0C1S0 A0A0P5NI37 A0A0P5JXZ9 A0A0P5PFI2 A0A0P5LY81 A0A3P8NEL8 A0A0P5J3H5 A0A0P5GF13 A0A0P5G933 A0A0N8BJY3 A0A0P6H3Y9 A0A0N8DXE3 A0A3P8NEN0 A0A0P5RWQ5 A0A0P5FXD2 A0A0N8AUY6 A0A0P5IV54 A0A0P5RZH8 A0A0P5PN39 A0A3P9AXL2 A0A0P5DXE5 A0A3P9AXR5 A0A0P5GPI4 A0A0P5L7M2 A0A0P6E3V7 A0A0P5K179 A0A3P8NET0 A0A0P5J1F9 A0A0P5SE30 A0A0P4Z186 A0A0P5I355 A0A0N8CGL0 A0A0P6DFB9 A0A0P5CR20 A0A0P5Z3A9 A0A0P5EGI9 A0A0P5KZS4 A0A0P6E9J7 A0A0P5K6Y3 A0A0P5Q0G8 A0A0N8C0M5 Q2YI44 A0A0P6A4V9 A0A3Q3BHZ1 A0A3B4FMG6 A0A0N8BKC7 A0A1B1UZF6 A0A1B6KIS9 A0A0P5KAT0 A0A023FWP6 A0A3B4BJ67 A0A0P5AT73 A0A0P5LKJ2 A0A0P5FUT7 A0A0P6GYI7 A0A0P6B1N1 A0A0P5LPV1 A0A0P5KGH2 A0A0P4YFW2 A0A0T6B1R3 A0A1B6JXC2 Q1T728 A0A1A9V729 Q45VP9 A0A2J7QQP4 E9H6F1 A0A1B6I2G9 B3S2H0 A0A0P5EP04 A0A369SSL3 A0A3Q2LBF6 A0A1I8MWR0 I3KFZ1 I3KFY9 S7P8B8 I3KFZ0
PDB
3M0C
E-value=4.62014e-28,
Score=310
Ontologies
GO
PANTHER
Topology
SignalP
Position: 1 - 17,
Likelihood: 0.999619
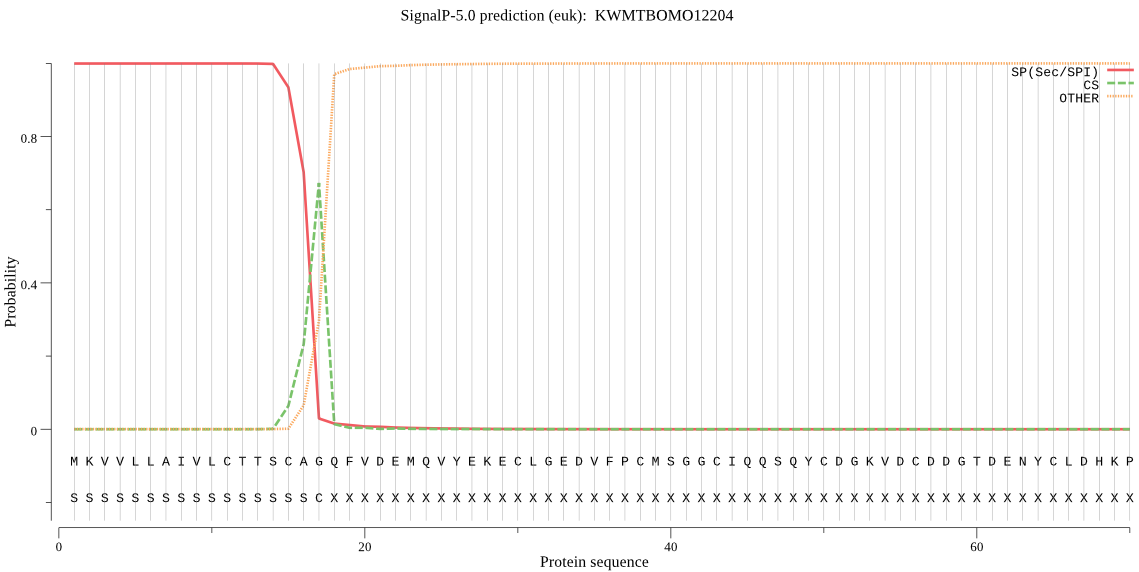
Length:
377
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.17005
Exp number, first 60 AAs:
0.16699
Total prob of N-in:
0.01188
outside
1 - 377

Population Genetic Test Statistics
Pi
258.306646
Theta
157.559306
Tajima's D
2.265754
CLR
10.342689
CSRT
0.92010399480026
Interpretation
Uncertain
