Gene
KWMTBOMO12201
Pre Gene Modal
BGIBMGA014163
Annotation
vitellogenin_receptor_[Bombyx_mori]
Full name
Putative vitellogenin receptor
+ More
Vitellogenin receptor
Vitellogenin receptor
Alternative Name
Protein yolkless
Location in the cell
Extracellular Reliability : 3.443
Sequence
CDS
ATGATGGGGTTCCCACCAAAGAGCGTATCGTGCAAGCCCGGGTACTATCAATGCCGCGACCGGGAGTGTATAGAGCTGAAGAAGCGCTGCGACGGACACCAGGACTGTTTTGATTACTCCGACGAGGAAGAGTGTGATGAGCCAGTTGTCGTGGAGGAGCCGAAAATACATCGTTGTGCCGAATGGGAGTACAGTTGCGAGCGTAACAGAAGTATCTGTTTACCGATTACGGCAAGGTGCAACATGAAAACCGACTGCCCTGGTGGAACGGATGAGATAGGCTGCGACTACCGGTGCACTCCTCACGGCATGTTCGGTTGCAAGCAGCAGATCCGGTGCTTGGCCATGAACCGGGTTTGCGACGGAAACAAGGAGTGCGACGATGGATCTGATGAGACGCCCGACGCTTGCGCTCTCGTCAACAGAACCTCCCACCTGTACCCGGTGATGCTGTATCCGGCAGCAGAGTGCCGCGACGGATTCCTCTGCGGCAACGGTCAATGCATCGAGTGGGCGGAAGTGTGCGACCGCACCCCCAACTGCTTCGACGGATCGGACGAGAGCATCCACTGCTTCTCGGCGTGCGACAACAACACGTGCGCCCACGCGTGCCAGGCCACGCCGCTGGGGCCGCGCTGCCTGTGTCCGGCCGGGTACAGCGCCGCGCCGGACCGCCGGACGTGCGCCGACGTGGACGAGTGCCGCGCGGGACTGTGCTCGCAGGCCTGCGTCAACACCCCCGGCTCCTTCCTCTGCTCGTGCCATCACGGGTACGCGCTTAGGTCCGACAGACGTTCTTGCAAGGCCGTCACCGGGAACATGTCCATACTGTACGTGTCTGGCAACACCGTGCGCTCCGTCTCGGCTGACGGCTACGGCGCCATAGAGTACAGCGACCCGGACCTTGGCGATATCACTGATTTGGATTTCAATGTTAGAACGAAGCGTTTGTATGTGACGTCTACGGAGTCGGGGAAGCTGATAGAATTGAACGTGACACATGACGTGGTCGCCGTGACGAACGTCGGACGGCCGACCAGGGTGGCAGTGGACTGGGTGACGGGCAACGTGTACTTCGCGGACAGCACGCCGGGTGCTAGCTGCGTGAGGGTCTGTGACGTCACCAGGAGGAGATGCGCCAGGCTGCAGAAGATACCCTCTGACGCAACGGTTAGTGCACGTCCAGAACTTGTTGTATACTGA
Protein
MMGFPPKSVSCKPGYYQCRDRECIELKKRCDGHQDCFDYSDEEECDEPVVVEEPKIHRCAEWEYSCERNRSICLPITARCNMKTDCPGGTDEIGCDYRCTPHGMFGCKQQIRCLAMNRVCDGNKECDDGSDETPDACALVNRTSHLYPVMLYPAAECRDGFLCGNGQCIEWAEVCDRTPNCFDGSDESIHCFSACDNNTCAHACQATPLGPRCLCPAGYSAAPDRRTCADVDECRAGLCSQACVNTPGSFLCSCHHGYALRSDRRSCKAVTGNMSILYVSGNTVRSVSADGYGAIEYSDPDLGDITDLDFNVRTKRLYVTSTESGKLIELNVTHDVVAVTNVGRPTRVAVDWVTGNVYFADSTPGASCVRVCDVTRRRCARLQKIPSDATVSARPELVVY
Summary
Description
Involved in uptake of vitellogenin by endocytosis. Expression is regulated by the juvenile hormone analog, methoprene (in vitro).
Keywords
Alternative splicing
Cell membrane
Complete proteome
Disulfide bond
EGF-like domain
Endocytosis
Glycoprotein
Membrane
Phosphoprotein
Receptor
Reference proteome
Repeat
Signal
Transmembrane
Transmembrane helix
Calcium
Feature
chain Putative vitellogenin receptor
splice variant In isoform A.
splice variant In isoform A.
Uniprot
Q66NE3
K4MUW4
Q66NE4
E2FLQ4
H2CLV8
H9JX98
+ More
A0A385LM65 A0A2A4JF17 A0A2D0VP24 M1L4G2 D9ZFI0 A0A0N1PFQ7 A0A194QLA3 A0A1Z2RRL4 A0A385HW27 A0A212FF10 A0A0T6B1R3 A0A2S1XV31 A0A1B6CX71 A0A1L8E338 A0A3G1KL05 X1WIS9 A0A1B1UZF6 A0A0B4ZZA6 B7QMR1 A0A2I4PYC0 K7IWA8 A0A139WE08 A0A232F2C2 Q7PS28 A0A0M4ETD4 A0A182JZY0 A0A2W1B456 A0A182UAS0 A0A2Y9D3I2 A0A293M1D3 A0A182IAT1 A0A182T7F8 A0A182RMV4 A0A0P8XJ28 A0A147BAB1 B3MZU7 A0A310S5G3 P98163-2 A0A0D4RRS9 P98163 A0A0L7QUY6 A0A182W5F8 A0A1S4FH50 A0A2A3EJ30 B4Q2Q9 A0A182XXV7 B3NVV7 E2BHT7 Q16GY3 O76952 D5KXW4 A0A182G7W8 B4IGA9 Q171G8 A0A1Y1N8E6 E0VAH7 A0A0M9A5Y8 Q2YI44 A0A151WPZ7 B1Q2W6 T1E9W2 W4VRI0 A0A087ZPZ1 A0A1W4VH45 A0A2J7QQP4 A0A084WEK1 A0A0Q9XEB7 A0A0C9QAX1 Q6X0I2 B4L5J5 B4JWV0 A0A151INQ8 A0A0Q9WBK4 B4M308 B4H0I5 Q29H17 Q8MP02 A0A0K8WJW4 A0A2I7G3Y1 A0A0K8U2F7 A0A131YJ60 U5KCA6 A0A3B5M1J7 A0A182PBK9 A0A182IR31 S4TMC4 A0A0S7FWJ3 A0A1W5KSB7 A0A1A9W8J7 A0A3B5ME55
A0A385LM65 A0A2A4JF17 A0A2D0VP24 M1L4G2 D9ZFI0 A0A0N1PFQ7 A0A194QLA3 A0A1Z2RRL4 A0A385HW27 A0A212FF10 A0A0T6B1R3 A0A2S1XV31 A0A1B6CX71 A0A1L8E338 A0A3G1KL05 X1WIS9 A0A1B1UZF6 A0A0B4ZZA6 B7QMR1 A0A2I4PYC0 K7IWA8 A0A139WE08 A0A232F2C2 Q7PS28 A0A0M4ETD4 A0A182JZY0 A0A2W1B456 A0A182UAS0 A0A2Y9D3I2 A0A293M1D3 A0A182IAT1 A0A182T7F8 A0A182RMV4 A0A0P8XJ28 A0A147BAB1 B3MZU7 A0A310S5G3 P98163-2 A0A0D4RRS9 P98163 A0A0L7QUY6 A0A182W5F8 A0A1S4FH50 A0A2A3EJ30 B4Q2Q9 A0A182XXV7 B3NVV7 E2BHT7 Q16GY3 O76952 D5KXW4 A0A182G7W8 B4IGA9 Q171G8 A0A1Y1N8E6 E0VAH7 A0A0M9A5Y8 Q2YI44 A0A151WPZ7 B1Q2W6 T1E9W2 W4VRI0 A0A087ZPZ1 A0A1W4VH45 A0A2J7QQP4 A0A084WEK1 A0A0Q9XEB7 A0A0C9QAX1 Q6X0I2 B4L5J5 B4JWV0 A0A151INQ8 A0A0Q9WBK4 B4M308 B4H0I5 Q29H17 Q8MP02 A0A0K8WJW4 A0A2I7G3Y1 A0A0K8U2F7 A0A131YJ60 U5KCA6 A0A3B5M1J7 A0A182PBK9 A0A182IR31 S4TMC4 A0A0S7FWJ3 A0A1W5KSB7 A0A1A9W8J7 A0A3B5ME55
Pubmed
23515308
19121390
20955241
26354079
28605547
22118469
+ More
29718485 27464714 20075255 18362917 19820115 28648823 12364791 14747013 17210077 28756777 17994087 7878005 10731132 12537572 18327897 17550304 25244985 20798317 17510324 8799131 11881811 26483478 28004739 20566863 16403020 24438588 15056367 15632085 16033432 29054547 26830274 24128609 26262609
29718485 27464714 20075255 18362917 19820115 28648823 12364791 14747013 17210077 28756777 17994087 7878005 10731132 12537572 18327897 17550304 25244985 20798317 17510324 8799131 11881811 26483478 28004739 20566863 16403020 24438588 15056367 15632085 16033432 29054547 26830274 24128609 26262609
EMBL
AY676608
AAU12845.1
JQ809472
AFV32171.1
AY676607
AAU12844.1
+ More
HM172611 ADK94452.1 JN003583 AEJ88360.1 BABH01035721 MH745217 AYA29178.2 NWSH01001818 PCG70030.1 KT899978 AOX13593.1 KC181922 AGF33811.2 GU983858 ADK94033.1 KQ461113 KPJ08910.1 KQ459053 KPJ04221.1 KY963162 ASA46482.1 MG799569 AXY55007.1 AGBW02008886 OWR52293.1 LJIG01016164 KRT81424.1 MF622539 AWJ95281.1 GEDC01019209 JAS18089.1 GFDF01000931 JAV13153.1 KY986532 ATU47286.1 ABLF02017882 ABLF02017885 ABLF02017888 ABLF02017889 KX428486 ANW09597.1 KM085006 AJD79559.1 ABJB011082826 DS972524 EEC20133.1 KY373243 AQS83398.1 KQ971355 KYB26208.1 NNAY01001168 OXU24904.1 AAAB01008846 EAA06264.6 CP012528 ALC49752.1 KZ150467 PZC70778.1 GFWV01009983 MAA34712.1 APCN01001317 APCN01001318 CH902635 KPU74830.1 GEIB01000012 JAR87707.1 EDV33898.1 KQ769958 OAD52717.1 U13637 AE014298 BT003478 BT150470 AJV26740.1 KQ414731 KOC62359.1 KZ288227 PBC31775.1 CM000162 EDX01653.2 CH954180 EDV47122.1 GL448324 EFN84770.1 CH478226 EAT33503.1 L77800 AH010501 AAC28497.1 AAK15810.1 GU723297 ADE34166.1 JXUM01046908 JXUM01046909 KQ561496 KXJ78386.1 CH480835 EDW48843.1 CH477456 EAT40623.1 GEZM01010121 JAV94182.1 DS235006 EEB10383.1 KQ435746 KOX76539.1 AM050637 CAJ19121.1 KQ982851 KYQ49878.1 AB299015 BAG14342.1 GAMD01001553 JAB00038.1 GANO01003098 JAB56773.1 NEVH01012083 PNF30911.1 ATLV01023220 KE525341 KFB48645.1 CH933811 KRG06924.1 GBYB01000374 JAG70141.1 AY262832 AAP92450.1 EDW06454.1 CH916376 EDV95226.1 KQ976914 KYN07112.1 CH940651 KRF82061.1 EDW65183.2 CH479200 EDW29780.1 CH379064 EAL31942.2 AB077047 BAC02725.2 GDHF01000878 JAI51436.1 KY781176 AUQ44344.1 GDHF01031628 JAI20686.1 GEDV01010025 JAP78532.1 JX846592 AGQ57038.1 JX469118 AGE83235.1 GBYX01460207 GBYX01460205 JAO21359.1 KR401221 AMZ04157.1
HM172611 ADK94452.1 JN003583 AEJ88360.1 BABH01035721 MH745217 AYA29178.2 NWSH01001818 PCG70030.1 KT899978 AOX13593.1 KC181922 AGF33811.2 GU983858 ADK94033.1 KQ461113 KPJ08910.1 KQ459053 KPJ04221.1 KY963162 ASA46482.1 MG799569 AXY55007.1 AGBW02008886 OWR52293.1 LJIG01016164 KRT81424.1 MF622539 AWJ95281.1 GEDC01019209 JAS18089.1 GFDF01000931 JAV13153.1 KY986532 ATU47286.1 ABLF02017882 ABLF02017885 ABLF02017888 ABLF02017889 KX428486 ANW09597.1 KM085006 AJD79559.1 ABJB011082826 DS972524 EEC20133.1 KY373243 AQS83398.1 KQ971355 KYB26208.1 NNAY01001168 OXU24904.1 AAAB01008846 EAA06264.6 CP012528 ALC49752.1 KZ150467 PZC70778.1 GFWV01009983 MAA34712.1 APCN01001317 APCN01001318 CH902635 KPU74830.1 GEIB01000012 JAR87707.1 EDV33898.1 KQ769958 OAD52717.1 U13637 AE014298 BT003478 BT150470 AJV26740.1 KQ414731 KOC62359.1 KZ288227 PBC31775.1 CM000162 EDX01653.2 CH954180 EDV47122.1 GL448324 EFN84770.1 CH478226 EAT33503.1 L77800 AH010501 AAC28497.1 AAK15810.1 GU723297 ADE34166.1 JXUM01046908 JXUM01046909 KQ561496 KXJ78386.1 CH480835 EDW48843.1 CH477456 EAT40623.1 GEZM01010121 JAV94182.1 DS235006 EEB10383.1 KQ435746 KOX76539.1 AM050637 CAJ19121.1 KQ982851 KYQ49878.1 AB299015 BAG14342.1 GAMD01001553 JAB00038.1 GANO01003098 JAB56773.1 NEVH01012083 PNF30911.1 ATLV01023220 KE525341 KFB48645.1 CH933811 KRG06924.1 GBYB01000374 JAG70141.1 AY262832 AAP92450.1 EDW06454.1 CH916376 EDV95226.1 KQ976914 KYN07112.1 CH940651 KRF82061.1 EDW65183.2 CH479200 EDW29780.1 CH379064 EAL31942.2 AB077047 BAC02725.2 GDHF01000878 JAI51436.1 KY781176 AUQ44344.1 GDHF01031628 JAI20686.1 GEDV01010025 JAP78532.1 JX846592 AGQ57038.1 JX469118 AGE83235.1 GBYX01460207 GBYX01460205 JAO21359.1 KR401221 AMZ04157.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000007151
UP000007819
+ More
UP000001555 UP000002358 UP000007266 UP000215335 UP000007062 UP000092553 UP000075881 UP000075902 UP000076407 UP000075840 UP000075901 UP000075900 UP000007801 UP000000803 UP000053825 UP000075920 UP000242457 UP000002282 UP000076408 UP000008711 UP000008237 UP000008820 UP000069940 UP000249989 UP000001292 UP000009046 UP000053105 UP000075809 UP000005203 UP000192221 UP000235965 UP000030765 UP000009192 UP000001070 UP000078542 UP000008792 UP000008744 UP000001819 UP000261380 UP000075885 UP000075880 UP000091820
UP000001555 UP000002358 UP000007266 UP000215335 UP000007062 UP000092553 UP000075881 UP000075902 UP000076407 UP000075840 UP000075901 UP000075900 UP000007801 UP000000803 UP000053825 UP000075920 UP000242457 UP000002282 UP000076408 UP000008711 UP000008237 UP000008820 UP000069940 UP000249989 UP000001292 UP000009046 UP000053105 UP000075809 UP000005203 UP000192221 UP000235965 UP000030765 UP000009192 UP000001070 UP000078542 UP000008792 UP000008744 UP000001819 UP000261380 UP000075885 UP000075880 UP000091820
Pfam
Interpro
IPR000152
EGF-type_Asp/Asn_hydroxyl_site
+ More
IPR023415 LDLR_class-A_CS
IPR018097 EGF_Ca-bd_CS
IPR013032 EGF-like_CS
IPR000742 EGF-like_dom
IPR040093 LRAD1
IPR011042 6-blade_b-propeller_TolB-like
IPR009030 Growth_fac_rcpt_cys_sf
IPR000033 LDLR_classB_rpt
IPR001881 EGF-like_Ca-bd_dom
IPR002172 LDrepeatLR_classA_rpt
IPR036055 LDL_receptor-like_sf
IPR026823 cEGF
IPR001190 SRCR
IPR006612 THAP_Znf
IPR003892 CUE
IPR019136 TF_IIIC_su-5_HTH
IPR041499 Tau95_N
IPR023415 LDLR_class-A_CS
IPR018097 EGF_Ca-bd_CS
IPR013032 EGF-like_CS
IPR000742 EGF-like_dom
IPR040093 LRAD1
IPR011042 6-blade_b-propeller_TolB-like
IPR009030 Growth_fac_rcpt_cys_sf
IPR000033 LDLR_classB_rpt
IPR001881 EGF-like_Ca-bd_dom
IPR002172 LDrepeatLR_classA_rpt
IPR036055 LDL_receptor-like_sf
IPR026823 cEGF
IPR001190 SRCR
IPR006612 THAP_Znf
IPR003892 CUE
IPR019136 TF_IIIC_su-5_HTH
IPR041499 Tau95_N
Gene 3D
CDD
ProteinModelPortal
Q66NE3
K4MUW4
Q66NE4
E2FLQ4
H2CLV8
H9JX98
+ More
A0A385LM65 A0A2A4JF17 A0A2D0VP24 M1L4G2 D9ZFI0 A0A0N1PFQ7 A0A194QLA3 A0A1Z2RRL4 A0A385HW27 A0A212FF10 A0A0T6B1R3 A0A2S1XV31 A0A1B6CX71 A0A1L8E338 A0A3G1KL05 X1WIS9 A0A1B1UZF6 A0A0B4ZZA6 B7QMR1 A0A2I4PYC0 K7IWA8 A0A139WE08 A0A232F2C2 Q7PS28 A0A0M4ETD4 A0A182JZY0 A0A2W1B456 A0A182UAS0 A0A2Y9D3I2 A0A293M1D3 A0A182IAT1 A0A182T7F8 A0A182RMV4 A0A0P8XJ28 A0A147BAB1 B3MZU7 A0A310S5G3 P98163-2 A0A0D4RRS9 P98163 A0A0L7QUY6 A0A182W5F8 A0A1S4FH50 A0A2A3EJ30 B4Q2Q9 A0A182XXV7 B3NVV7 E2BHT7 Q16GY3 O76952 D5KXW4 A0A182G7W8 B4IGA9 Q171G8 A0A1Y1N8E6 E0VAH7 A0A0M9A5Y8 Q2YI44 A0A151WPZ7 B1Q2W6 T1E9W2 W4VRI0 A0A087ZPZ1 A0A1W4VH45 A0A2J7QQP4 A0A084WEK1 A0A0Q9XEB7 A0A0C9QAX1 Q6X0I2 B4L5J5 B4JWV0 A0A151INQ8 A0A0Q9WBK4 B4M308 B4H0I5 Q29H17 Q8MP02 A0A0K8WJW4 A0A2I7G3Y1 A0A0K8U2F7 A0A131YJ60 U5KCA6 A0A3B5M1J7 A0A182PBK9 A0A182IR31 S4TMC4 A0A0S7FWJ3 A0A1W5KSB7 A0A1A9W8J7 A0A3B5ME55
A0A385LM65 A0A2A4JF17 A0A2D0VP24 M1L4G2 D9ZFI0 A0A0N1PFQ7 A0A194QLA3 A0A1Z2RRL4 A0A385HW27 A0A212FF10 A0A0T6B1R3 A0A2S1XV31 A0A1B6CX71 A0A1L8E338 A0A3G1KL05 X1WIS9 A0A1B1UZF6 A0A0B4ZZA6 B7QMR1 A0A2I4PYC0 K7IWA8 A0A139WE08 A0A232F2C2 Q7PS28 A0A0M4ETD4 A0A182JZY0 A0A2W1B456 A0A182UAS0 A0A2Y9D3I2 A0A293M1D3 A0A182IAT1 A0A182T7F8 A0A182RMV4 A0A0P8XJ28 A0A147BAB1 B3MZU7 A0A310S5G3 P98163-2 A0A0D4RRS9 P98163 A0A0L7QUY6 A0A182W5F8 A0A1S4FH50 A0A2A3EJ30 B4Q2Q9 A0A182XXV7 B3NVV7 E2BHT7 Q16GY3 O76952 D5KXW4 A0A182G7W8 B4IGA9 Q171G8 A0A1Y1N8E6 E0VAH7 A0A0M9A5Y8 Q2YI44 A0A151WPZ7 B1Q2W6 T1E9W2 W4VRI0 A0A087ZPZ1 A0A1W4VH45 A0A2J7QQP4 A0A084WEK1 A0A0Q9XEB7 A0A0C9QAX1 Q6X0I2 B4L5J5 B4JWV0 A0A151INQ8 A0A0Q9WBK4 B4M308 B4H0I5 Q29H17 Q8MP02 A0A0K8WJW4 A0A2I7G3Y1 A0A0K8U2F7 A0A131YJ60 U5KCA6 A0A3B5M1J7 A0A182PBK9 A0A182IR31 S4TMC4 A0A0S7FWJ3 A0A1W5KSB7 A0A1A9W8J7 A0A3B5ME55
PDB
5B4X
E-value=7.23356e-33,
Score=351
Ontologies
GO
PANTHER
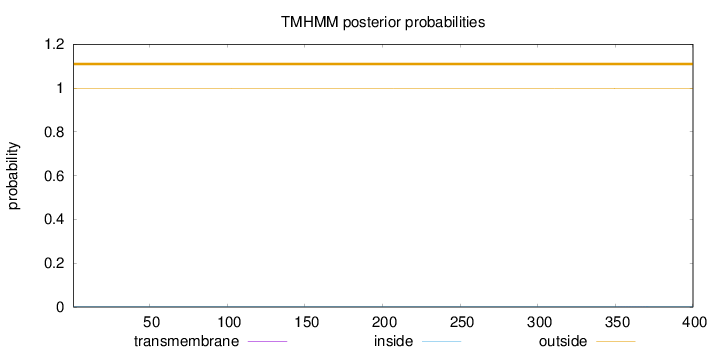
Topology
Subcellular location
Cell membrane
Membrane
Membrane
Length:
400
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00497
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00166
outside
1 - 400
Population Genetic Test Statistics
Pi
350.019948
Theta
190.629946
Tajima's D
2.977596
CLR
0.103877
CSRT
0.977601119944003
Interpretation
Uncertain
