Gene
KWMTBOMO12196
Pre Gene Modal
BGIBMGA014176
Annotation
Transcriptional_repressor_scratch_1_[Papilio_xuthus]
Transcription factor
Location in the cell
Mitochondrial Reliability : 1.475 Nuclear Reliability : 2.027
Sequence
CDS
ATGCACAAATGCAAGTATTGCGGGAAAGGCTTCGACAGACCCTGGGTGCTGAAGGGTCACCTGCGTCTCCACACCGGGGAGAGACCCTTCCCGTGCCCCCATCCGCTCTGCGGCAGAAGCTTTGCTGATCGTTCGAACCTCCGCGCCCACCAGCGCACCCGCGGTCACCACTCGTGGCAGTGGCGCTGCTCGGAGTGTGGGAAAGCTTTCAGCCAGAGCCGCTACCTCGAGCGCCACCGAGAGGACGCTTGTCGAAAATATAAGTACGTTGCGTTCGATGTTAAGTTCTAG
Protein
MHKCKYCGKGFDRPWVLKGHLRLHTGERPFPCPHPLCGRSFADRSNLRAHQRTRGHHSWQWRCSECGKAFSQSRYLERHREDACRKYKYVAFDVKF
Summary
Uniprot
H9JXB1
A0A194QL98
A0A2A4JE19
A0A0N1PH24
A0A3S2M3Y2
A0A212FEX1
+ More
A0A2H1VE54 A0A2P8XUU1 A0A0L7QUQ6 A0A087ZPS1 A0A154P013 A0A0L7KTL9 A0A0N0BHQ0 A0A232F3A2 E0VJ65 K7IWA7 A0A0J7KU98 E2AXW5 A0A151INT4 A0A195EVR1 A0A026WJK6 A0A158NXN4 A0A195B0C3 E0W044 F4WYD0 A0A195EDX2 A0A2A3EJX0 E9J0P6 A0A0C9PKZ9 A0A1B6GZ14 A0A1B6KPN9 A0A0L0BVT2 A0A1A9V606 A0A1I8NWL1 A0A1I8MN07 A0A1A9Y9F2 A0A1A9W2R9 A0A1B0F9W1 W8BX65 A0A1B0C7M6 A0A0K8UEL3 A0A0A1X3X8 A0A034WS72 A0A1A9ZE94 Q299M7 B4G5F6 B4IUZ0 B3NZ50 B4NH31 A0A3B0K5P4 A0A0M4ELT3 B4I282 B4M3Y5 A0A1W4UKY0 Q9VE63 B4K917 B3LW90 B4QUD9 B4JF08 T1IA04 A0A1I8I524 A0A336KKG5 Q95VQ7 T1FNI8 A0A087T657 A0A3B4DCX3 A0A3B1IHX9 U3JT54 T1KC02 A0A1V9XC04 U3IW47 H2TXK9 T1IYF8 A0A087UIF5 M7B4X2 W5M2K9 O16139 A0A226NCM6 A0A3L7HE06 A0A0V1MD77 A0A1X8XKV4 Q6UCK0 W4XCK8 Q0Z887 R7THH0 A0A1L8FU78 A0A0V1HYJ2 U3J8I8 A0A0V1ISU4 A0A0V1JT95 A0A0N5DM67 A0A0P7XBU3 A0A315UUZ7 I3KUZ1 A0A0V1FJP0 A0A3B3QG56 A0A3B3X4F5 A0A087XI97 A0A3B3U8E0 A0A3P9P7Z5
A0A2H1VE54 A0A2P8XUU1 A0A0L7QUQ6 A0A087ZPS1 A0A154P013 A0A0L7KTL9 A0A0N0BHQ0 A0A232F3A2 E0VJ65 K7IWA7 A0A0J7KU98 E2AXW5 A0A151INT4 A0A195EVR1 A0A026WJK6 A0A158NXN4 A0A195B0C3 E0W044 F4WYD0 A0A195EDX2 A0A2A3EJX0 E9J0P6 A0A0C9PKZ9 A0A1B6GZ14 A0A1B6KPN9 A0A0L0BVT2 A0A1A9V606 A0A1I8NWL1 A0A1I8MN07 A0A1A9Y9F2 A0A1A9W2R9 A0A1B0F9W1 W8BX65 A0A1B0C7M6 A0A0K8UEL3 A0A0A1X3X8 A0A034WS72 A0A1A9ZE94 Q299M7 B4G5F6 B4IUZ0 B3NZ50 B4NH31 A0A3B0K5P4 A0A0M4ELT3 B4I282 B4M3Y5 A0A1W4UKY0 Q9VE63 B4K917 B3LW90 B4QUD9 B4JF08 T1IA04 A0A1I8I524 A0A336KKG5 Q95VQ7 T1FNI8 A0A087T657 A0A3B4DCX3 A0A3B1IHX9 U3JT54 T1KC02 A0A1V9XC04 U3IW47 H2TXK9 T1IYF8 A0A087UIF5 M7B4X2 W5M2K9 O16139 A0A226NCM6 A0A3L7HE06 A0A0V1MD77 A0A1X8XKV4 Q6UCK0 W4XCK8 Q0Z887 R7THH0 A0A1L8FU78 A0A0V1HYJ2 U3J8I8 A0A0V1ISU4 A0A0V1JT95 A0A0N5DM67 A0A0P7XBU3 A0A315UUZ7 I3KUZ1 A0A0V1FJP0 A0A3B3QG56 A0A3B3X4F5 A0A087XI97 A0A3B3U8E0 A0A3P9P7Z5
Pubmed
19121390
26354079
22118469
29403074
26227816
28648823
+ More
20566863 20075255 20798317 24508170 30249741 21347285 21719571 21282665 26108605 25315136 24495485 25830018 25348373 15632085 17994087 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26392545 11466529 23254933 25329095 28327890 21551351 23624526 24621616 29704459 28436008 17287249 27762356 29703783 25186727 29240929
20566863 20075255 20798317 24508170 30249741 21347285 21719571 21282665 26108605 25315136 24495485 25830018 25348373 15632085 17994087 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26392545 11466529 23254933 25329095 28327890 21551351 23624526 24621616 29704459 28436008 17287249 27762356 29703783 25186727 29240929
EMBL
BABH01035714
KQ459053
KPJ04216.1
NWSH01001818
PCG70026.1
KQ461113
+ More
KPJ08905.1 RSAL01000042 RVE50793.1 AGBW02008886 OWR52296.1 ODYU01002071 SOQ39118.1 PYGN01001311 PSN35781.1 KQ414731 KOC62358.1 KQ434782 KZC04588.1 JTDY01005799 KOB66612.1 KQ435746 KOX76538.1 NNAY01001168 OXU24907.1 DS235219 EEB13421.1 LBMM01003169 KMQ93866.1 GL443736 EFN61729.1 KQ976914 KYN07111.1 KQ981958 KYN31987.1 KK107168 QOIP01000001 EZA56180.1 RLU26775.1 ADTU01003248 KQ976692 KYM77740.1 DS235855 EEB19000.1 GL888440 EGI60782.1 KQ979074 KYN23002.1 KZ288227 PBC31774.1 GL767538 EFZ13544.1 GBYB01001678 JAG71445.1 GECZ01002099 JAS67670.1 GEBQ01026559 JAT13418.1 JRES01001362 KNC23349.1 CCAG010014901 GAMC01002698 JAC03858.1 JXJN01029245 JXJN01029246 GDHF01027220 JAI25094.1 GBXI01008939 JAD05353.1 GAKP01001790 JAC57162.1 CM000070 EAL27676.1 CH479179 EDW24822.1 CM000160 CH892279 EDW95918.1 EDX00204.1 CH954181 EDV48592.1 CH964272 EDW84528.1 OUUW01000013 SPP87992.1 CP012526 ALC46315.1 CH480820 EDW54639.1 CH940652 EDW59346.1 AE014297 BT022138 AAF55566.1 AAY51533.1 CH933806 EDW14430.1 CH902617 EDV42668.2 CM000364 EDX12439.1 CH916369 EDV93289.1 ACPB03005149 UFQS01000563 UFQT01000563 SSX04979.1 SSX25341.1 AF410865 AAL05565.1 AMQM01006851 KB097528 ESN95392.1 KK113612 KFM60596.1 AGTO01021867 CAEY01001956 MNPL01015858 OQR70882.1 JH431683 KK119951 KFM77144.1 KB572999 EMP27188.1 AHAT01021144 AHAT01021145 AF013243 AAB67715.1 AWGT02005927 OXB65020.1 RAZU01000234 RLQ64177.1 JYDO01000134 KRZ69539.1 LT821716 SLL96481.1 AY372519 AAQ74988.1 AAGJ04029712 DQ665364 ABG56163.1 AMQN01002676 KB309853 ELT93164.1 CM004477 OCT75152.1 JYDP01000017 KRZ15537.1 JYDU01000135 JYDS01000094 KRX91542.1 KRZ25858.1 JYDR01000111 JYDV01000053 KRY68457.1 KRZ37797.1 JARO02002006 KPP73854.1 NHOQ01002733 PWA15231.1 AERX01028453 JYDT01000076 KRY86178.1 AYCK01010451
KPJ08905.1 RSAL01000042 RVE50793.1 AGBW02008886 OWR52296.1 ODYU01002071 SOQ39118.1 PYGN01001311 PSN35781.1 KQ414731 KOC62358.1 KQ434782 KZC04588.1 JTDY01005799 KOB66612.1 KQ435746 KOX76538.1 NNAY01001168 OXU24907.1 DS235219 EEB13421.1 LBMM01003169 KMQ93866.1 GL443736 EFN61729.1 KQ976914 KYN07111.1 KQ981958 KYN31987.1 KK107168 QOIP01000001 EZA56180.1 RLU26775.1 ADTU01003248 KQ976692 KYM77740.1 DS235855 EEB19000.1 GL888440 EGI60782.1 KQ979074 KYN23002.1 KZ288227 PBC31774.1 GL767538 EFZ13544.1 GBYB01001678 JAG71445.1 GECZ01002099 JAS67670.1 GEBQ01026559 JAT13418.1 JRES01001362 KNC23349.1 CCAG010014901 GAMC01002698 JAC03858.1 JXJN01029245 JXJN01029246 GDHF01027220 JAI25094.1 GBXI01008939 JAD05353.1 GAKP01001790 JAC57162.1 CM000070 EAL27676.1 CH479179 EDW24822.1 CM000160 CH892279 EDW95918.1 EDX00204.1 CH954181 EDV48592.1 CH964272 EDW84528.1 OUUW01000013 SPP87992.1 CP012526 ALC46315.1 CH480820 EDW54639.1 CH940652 EDW59346.1 AE014297 BT022138 AAF55566.1 AAY51533.1 CH933806 EDW14430.1 CH902617 EDV42668.2 CM000364 EDX12439.1 CH916369 EDV93289.1 ACPB03005149 UFQS01000563 UFQT01000563 SSX04979.1 SSX25341.1 AF410865 AAL05565.1 AMQM01006851 KB097528 ESN95392.1 KK113612 KFM60596.1 AGTO01021867 CAEY01001956 MNPL01015858 OQR70882.1 JH431683 KK119951 KFM77144.1 KB572999 EMP27188.1 AHAT01021144 AHAT01021145 AF013243 AAB67715.1 AWGT02005927 OXB65020.1 RAZU01000234 RLQ64177.1 JYDO01000134 KRZ69539.1 LT821716 SLL96481.1 AY372519 AAQ74988.1 AAGJ04029712 DQ665364 ABG56163.1 AMQN01002676 KB309853 ELT93164.1 CM004477 OCT75152.1 JYDP01000017 KRZ15537.1 JYDU01000135 JYDS01000094 KRX91542.1 KRZ25858.1 JYDR01000111 JYDV01000053 KRY68457.1 KRZ37797.1 JARO02002006 KPP73854.1 NHOQ01002733 PWA15231.1 AERX01028453 JYDT01000076 KRY86178.1 AYCK01010451
Proteomes
UP000005204
UP000053268
UP000218220
UP000053240
UP000283053
UP000007151
+ More
UP000245037 UP000053825 UP000005203 UP000076502 UP000037510 UP000053105 UP000215335 UP000009046 UP000002358 UP000036403 UP000000311 UP000078542 UP000078541 UP000053097 UP000279307 UP000005205 UP000078540 UP000007755 UP000078492 UP000242457 UP000037069 UP000078200 UP000095300 UP000095301 UP000092443 UP000091820 UP000092444 UP000092460 UP000092445 UP000001819 UP000008744 UP000002282 UP000008711 UP000007798 UP000268350 UP000092553 UP000001292 UP000008792 UP000192221 UP000000803 UP000009192 UP000007801 UP000000304 UP000001070 UP000015103 UP000095280 UP000015101 UP000054359 UP000261440 UP000018467 UP000016665 UP000015104 UP000192247 UP000005226 UP000031443 UP000018468 UP000198419 UP000273346 UP000054843 UP000007110 UP000014760 UP000186698 UP000055024 UP000054805 UP000054815 UP000054632 UP000054826 UP000046395 UP000034805 UP000005207 UP000054995 UP000261540 UP000261480 UP000028760 UP000261500 UP000242638
UP000245037 UP000053825 UP000005203 UP000076502 UP000037510 UP000053105 UP000215335 UP000009046 UP000002358 UP000036403 UP000000311 UP000078542 UP000078541 UP000053097 UP000279307 UP000005205 UP000078540 UP000007755 UP000078492 UP000242457 UP000037069 UP000078200 UP000095300 UP000095301 UP000092443 UP000091820 UP000092444 UP000092460 UP000092445 UP000001819 UP000008744 UP000002282 UP000008711 UP000007798 UP000268350 UP000092553 UP000001292 UP000008792 UP000192221 UP000000803 UP000009192 UP000007801 UP000000304 UP000001070 UP000015103 UP000095280 UP000015101 UP000054359 UP000261440 UP000018467 UP000016665 UP000015104 UP000192247 UP000005226 UP000031443 UP000018468 UP000198419 UP000273346 UP000054843 UP000007110 UP000014760 UP000186698 UP000055024 UP000054805 UP000054815 UP000054632 UP000054826 UP000046395 UP000034805 UP000005207 UP000054995 UP000261540 UP000261480 UP000028760 UP000261500 UP000242638
Interpro
SUPFAM
SSF57667
SSF57667
ProteinModelPortal
H9JXB1
A0A194QL98
A0A2A4JE19
A0A0N1PH24
A0A3S2M3Y2
A0A212FEX1
+ More
A0A2H1VE54 A0A2P8XUU1 A0A0L7QUQ6 A0A087ZPS1 A0A154P013 A0A0L7KTL9 A0A0N0BHQ0 A0A232F3A2 E0VJ65 K7IWA7 A0A0J7KU98 E2AXW5 A0A151INT4 A0A195EVR1 A0A026WJK6 A0A158NXN4 A0A195B0C3 E0W044 F4WYD0 A0A195EDX2 A0A2A3EJX0 E9J0P6 A0A0C9PKZ9 A0A1B6GZ14 A0A1B6KPN9 A0A0L0BVT2 A0A1A9V606 A0A1I8NWL1 A0A1I8MN07 A0A1A9Y9F2 A0A1A9W2R9 A0A1B0F9W1 W8BX65 A0A1B0C7M6 A0A0K8UEL3 A0A0A1X3X8 A0A034WS72 A0A1A9ZE94 Q299M7 B4G5F6 B4IUZ0 B3NZ50 B4NH31 A0A3B0K5P4 A0A0M4ELT3 B4I282 B4M3Y5 A0A1W4UKY0 Q9VE63 B4K917 B3LW90 B4QUD9 B4JF08 T1IA04 A0A1I8I524 A0A336KKG5 Q95VQ7 T1FNI8 A0A087T657 A0A3B4DCX3 A0A3B1IHX9 U3JT54 T1KC02 A0A1V9XC04 U3IW47 H2TXK9 T1IYF8 A0A087UIF5 M7B4X2 W5M2K9 O16139 A0A226NCM6 A0A3L7HE06 A0A0V1MD77 A0A1X8XKV4 Q6UCK0 W4XCK8 Q0Z887 R7THH0 A0A1L8FU78 A0A0V1HYJ2 U3J8I8 A0A0V1ISU4 A0A0V1JT95 A0A0N5DM67 A0A0P7XBU3 A0A315UUZ7 I3KUZ1 A0A0V1FJP0 A0A3B3QG56 A0A3B3X4F5 A0A087XI97 A0A3B3U8E0 A0A3P9P7Z5
A0A2H1VE54 A0A2P8XUU1 A0A0L7QUQ6 A0A087ZPS1 A0A154P013 A0A0L7KTL9 A0A0N0BHQ0 A0A232F3A2 E0VJ65 K7IWA7 A0A0J7KU98 E2AXW5 A0A151INT4 A0A195EVR1 A0A026WJK6 A0A158NXN4 A0A195B0C3 E0W044 F4WYD0 A0A195EDX2 A0A2A3EJX0 E9J0P6 A0A0C9PKZ9 A0A1B6GZ14 A0A1B6KPN9 A0A0L0BVT2 A0A1A9V606 A0A1I8NWL1 A0A1I8MN07 A0A1A9Y9F2 A0A1A9W2R9 A0A1B0F9W1 W8BX65 A0A1B0C7M6 A0A0K8UEL3 A0A0A1X3X8 A0A034WS72 A0A1A9ZE94 Q299M7 B4G5F6 B4IUZ0 B3NZ50 B4NH31 A0A3B0K5P4 A0A0M4ELT3 B4I282 B4M3Y5 A0A1W4UKY0 Q9VE63 B4K917 B3LW90 B4QUD9 B4JF08 T1IA04 A0A1I8I524 A0A336KKG5 Q95VQ7 T1FNI8 A0A087T657 A0A3B4DCX3 A0A3B1IHX9 U3JT54 T1KC02 A0A1V9XC04 U3IW47 H2TXK9 T1IYF8 A0A087UIF5 M7B4X2 W5M2K9 O16139 A0A226NCM6 A0A3L7HE06 A0A0V1MD77 A0A1X8XKV4 Q6UCK0 W4XCK8 Q0Z887 R7THH0 A0A1L8FU78 A0A0V1HYJ2 U3J8I8 A0A0V1ISU4 A0A0V1JT95 A0A0N5DM67 A0A0P7XBU3 A0A315UUZ7 I3KUZ1 A0A0V1FJP0 A0A3B3QG56 A0A3B3X4F5 A0A087XI97 A0A3B3U8E0 A0A3P9P7Z5
PDB
3W5K
E-value=4.15117e-19,
Score=226
Ontologies
GO
PANTHER
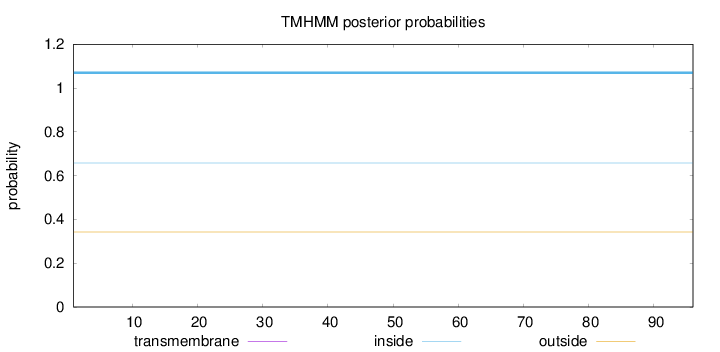
Topology
Length:
96
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.65737
inside
1 - 96
Population Genetic Test Statistics
Pi
348.30717
Theta
187.686928
Tajima's D
2.693039
CLR
0.152333
CSRT
0.96025198740063
Interpretation
Uncertain
