Gene
KWMTBOMO12195
Pre Gene Modal
BGIBMGA014169
Annotation
PREDICTED:_uncharacterized_protein_LOC106135765_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 1.032 Nuclear Reliability : 1.228
Sequence
CDS
ATGTGGTACACGGTGAGCGGTTCCGCCGAGTTCGTGTGCCCTATGAGATCGAATTGCTGCTCGCGGAGGTCGCCCTCGTATCGCGTGACGAAACCGTCGGTGGACGCGACCGCCGAGCCGCCTCCCCAGCCGGAGCAGCATCTGTTGGAGCCCACGCCCTCAGTCCGAGACACGAAGCCGCAGTTCAAAGGACCGCGTCCGACAGGATGCAGTGTTTCAGATTCTATATCGCTATCTTCGCCGGTGGCATGCGCGTCCCCAAAAGAGGACGTGGAGGGTCCACCGATGCCGGCCAATGTCGCCACTCTGCCCGAAGAACACAGGTCCTGCCTCATGGGGAGAGTGGCTGGACAGCGACCCCGGAGGCGACGGGCAATGCTCATCTACGTGTGCGCTGCTGACTCCCAAGATTGCTGTGCGGAGAAGGGCGCTCTGCAGTGCGGTGCCGCTGCTAGACTCCGGGTGAGGGCCCGCAAGAGAGGATGGAGAGTCCACGTGGCAGATCTCCACTGGCGCTCGCCTCTGGAACAGCAGAGAGACCATCGGTTCCCGGTGCTATGCATAACTGAACTTACGCGTCAATCAGAGTTAGGCGCTGTGGTCCCCGTGCTATTCCTCAACTCAGGGCTGGGCACGCCGCTGCTGCCCCACACACTGGAGTGCGCAGACTTCAAGGCCGCCCTGGAGGCAGCTGATGACGAAAACGACAAGGCACTGCTGAACAAATGGTATACGCTGGACTGCAGCACCACGCCGCCGTGCTACCGCCTGGTACGAGGGGCTCCGGCGCGCTGGAGGCGGGAGATGGCGCGGACCCTCAACGTCCTCGTCCGGACCTTACCTCAAGAAGCGTGCGACGCCTACTTAACCACCGTGGTTGAACAGGCAAGTCACCAGACTTTCATTATGGAATTCGTTATGTCATTAGAACTGATAATAGAGGTGCAGCACACAGTGGTGATGAGCGTGGAGGCGGCGCGGCGCTGCGTGTGGGTGTGCCGGGCGGGGGGCGCGGGCGGAGGCGGGGTGGGGGCAGGGGTGGGGGGCGAGCCCGACGACGCCGCGCAGATCCACGAGCTGCAGCGCAGACTCGCCAACTTGCAGAAAGAGTTAAAGGTCCATCTAAGCGAGCGTAACATCATCCGAGCGAGCGTTCGCGGCCGTTCGGCGGCGAGCGGCGACGACGAGTACGCGGAGGGCGTGCGCGCGGCGCTCGTGGAGCGGCTGGAGGCGCTGCTGGACGCGCTGGTCTCCGAGCACGACGCGCCCCCGGGATACAACGGGATACCCACCAGCTTGTTAGACGAAATCAATGAACACCTTATATTCTGTCAGAAGTCGGCGCAATGTACTTCAAATAGGGAGATCACGCTGAACGAACTTAAAACATATATAACGGGAGACAGCAACGTGCCTCTGGCGCTGATAGGCGGCGCCGGCTGCGGTAAGGCGACCCTGGTGGCCCGCGCCGTCACCATGGCGCCGACCTGCCAACAAGACCTCGCTGTTATCATAAGATTTGTGGGTCTGACGTCCCAGTCCAGCAGCTCTCATCAGCTCCTCAAGTCCATAGTCGACCAGTGCCATGTCCTCAACACGGGCACGGTGTACAGAGGACGCAATGACGTGGCCTCGTTGTCCACTCTCCTCCCGACCCTGCTGACGTCGCTGGGTCGGAGACGGCCGGTGCTGCTGGCCATTGTAGGGGCGGACGCCCTGAGGGCCGCCCGGTGGCTTCCCGAACGATTGCCTGATAACGTTAAGATGATCGTCACTGTTACTGAAGAAAAAGATGAACCGTCGAGTTCAGCTATCATGGCTGTTCTATCGTCGGAGGACGGGCCCAAGGTGCTGAGGGCGCCGCCGGTGGGAGCTGAAGAAGCGAAAGCGGTGCTGACGGCGTGCGCCGCCACGTACAGCAAGACCGGGGCCGCCTCTCCGCCGGAGGCAGCCGTCAAGGCTGTGCAAAAATGCACCTTACCTCTATACGCGAAGATCCTGGCTTGGCAGATATCCTTATCGGCGGAAACGTTCACCGAACAAGAACCGGACGCGACTCCCGAGCTGGTCATCTGCGAGGACCTGGAGGGACAGTTGGTGCAGATCTTGGTGGCGACTGAGGCGGCGCTGGGTGCCGAGCGAGTGAAGAACTGCCTCGCTCTGCTGACGGCCTCCAGGCGGGGCTTGGACGACACCGAGATCTTGGACATGTTGGCGCATGACGAATTGTTCAGAAGCCATGAAACTTATTTGCCGTGGGCGCCGGCGTCCCTGTTCTGGCCGCAGCTGGCGGCGCTGCTGGCGCCGTGGGTGCGGTGGGACGCGCGGGGGGCGGCGCGCTGGCGGGACGCCGCGCTGGCCGGCGCCGCCCGGGACCGCTACCTGGCCGACGCCGAACACGACGTCAGGAGGACCCTCGTCGAGTACTACGAGGTGAGGTTCAGCCAACATGGCGGCCACTGCGGGCTGGTGTTTAGATGA
Protein
MWYTVSGSAEFVCPMRSNCCSRRSPSYRVTKPSVDATAEPPPQPEQHLLEPTPSVRDTKPQFKGPRPTGCSVSDSISLSSPVACASPKEDVEGPPMPANVATLPEEHRSCLMGRVAGQRPRRRRAMLIYVCAADSQDCCAEKGALQCGAAARLRVRARKRGWRVHVADLHWRSPLEQQRDHRFPVLCITELTRQSELGAVVPVLFLNSGLGTPLLPHTLECADFKAALEAADDENDKALLNKWYTLDCSTTPPCYRLVRGAPARWRREMARTLNVLVRTLPQEACDAYLTTVVEQASHQTFIMEFVMSLELIIEVQHTVVMSVEAARRCVWVCRAGGAGGGGVGAGVGGEPDDAAQIHELQRRLANLQKELKVHLSERNIIRASVRGRSAASGDDEYAEGVRAALVERLEALLDALVSEHDAPPGYNGIPTSLLDEINEHLIFCQKSAQCTSNREITLNELKTYITGDSNVPLALIGGAGCGKATLVARAVTMAPTCQQDLAVIIRFVGLTSQSSSSHQLLKSIVDQCHVLNTGTVYRGRNDVASLSTLLPTLLTSLGRRRPVLLAIVGADALRAARWLPERLPDNVKMIVTVTEEKDEPSSSAIMAVLSSEDGPKVLRAPPVGAEEAKAVLTACAATYSKTGAASPPEAAVKAVQKCTLPLYAKILAWQISLSAETFTEQEPDATPELVICEDLEGQLVQILVATEAALGAERVKNCLALLTASRRGLDDTEILDMLAHDELFRSHETYLPWAPASLFWPQLAALLAPWVRWDARGAARWRDAALAGAARDRYLADAEHDVRRTLVEYYEVRFSQHGGHCGLVFR
Summary
Uniprot
A0A2H1V5G8
A0A2A4JAS8
A0A212FLL4
A0A194QH02
A0A194QW14
A0A3S2NGX7
+ More
A0A1W4WH42 A0A1W4W6Q0 A0A1W4WID3 A0A1Y1LH95 D6WR21 A0A1Y1LF63 A0A2J7QAW1 U4UKL9 A0A1B6KJV3 N6U1N5 E0VII1 A0A1D2MLL7 A0A224XGX4 A0A226F4Y7 A0A146LA42 A0A1J1HNX5 A0A0K8TLW2 A0A182GL42 A0A1S4FLV2 A0A0P6ISD2 A0A1S4GW40 A0A182IED3 A0A182L2N3 A0A182X1F3 Q16WD7 A0A182K0E6 A0A0N7ZWD5 A0A0A1WRS3 B0W0A1 A0A0A1WXR7 A0A0P5Z4E0 A0A0P5L1U9 W8ANI1 A0A0P5GPH2 A0A0P5UFZ3 A0A0K8U7S4 A0A0P5LI65 A0A0P5D2C0 A0A0P6H1F6 A0A0P6B7Z9 A0A0P5IK15 A0A0P5NAM8 A0A0P6FFP5 A0A0P5NRK6 A0A0P5PUA0 A0A0P6DBI9 A0A0N8BYE9 A0A0P5DWT5 A0A0N8BPQ1 A0A0P5A522 A0A0P5FRE4 A0A164KBN3 A0A0P5N8V9 A0A0K8UFP0 A0A0K8VCE5 A0A182W7M3 A0A182F1B4 A0A034W167 A0A182RU62 A0A182MPC5 A0A0P5R2V8 A0A0P5WAA1 A0A182XZQ7 A0A182PJR6 A0A1A9Z411 B3MN52 A0A182IS92 B4JPE3 W5JEP8 B4M900 A0A182QHQ1 A0A084W7L3
A0A1W4WH42 A0A1W4W6Q0 A0A1W4WID3 A0A1Y1LH95 D6WR21 A0A1Y1LF63 A0A2J7QAW1 U4UKL9 A0A1B6KJV3 N6U1N5 E0VII1 A0A1D2MLL7 A0A224XGX4 A0A226F4Y7 A0A146LA42 A0A1J1HNX5 A0A0K8TLW2 A0A182GL42 A0A1S4FLV2 A0A0P6ISD2 A0A1S4GW40 A0A182IED3 A0A182L2N3 A0A182X1F3 Q16WD7 A0A182K0E6 A0A0N7ZWD5 A0A0A1WRS3 B0W0A1 A0A0A1WXR7 A0A0P5Z4E0 A0A0P5L1U9 W8ANI1 A0A0P5GPH2 A0A0P5UFZ3 A0A0K8U7S4 A0A0P5LI65 A0A0P5D2C0 A0A0P6H1F6 A0A0P6B7Z9 A0A0P5IK15 A0A0P5NAM8 A0A0P6FFP5 A0A0P5NRK6 A0A0P5PUA0 A0A0P6DBI9 A0A0N8BYE9 A0A0P5DWT5 A0A0N8BPQ1 A0A0P5A522 A0A0P5FRE4 A0A164KBN3 A0A0P5N8V9 A0A0K8UFP0 A0A0K8VCE5 A0A182W7M3 A0A182F1B4 A0A034W167 A0A182RU62 A0A182MPC5 A0A0P5R2V8 A0A0P5WAA1 A0A182XZQ7 A0A182PJR6 A0A1A9Z411 B3MN52 A0A182IS92 B4JPE3 W5JEP8 B4M900 A0A182QHQ1 A0A084W7L3
Pubmed
EMBL
ODYU01000783
SOQ36095.1
NWSH01002091
PCG69217.1
AGBW02007739
OWR54634.1
+ More
KQ459053 KPJ04215.1 KQ461181 KPJ07756.1 RSAL01000042 RVE50792.1 GEZM01057545 JAV72261.1 KQ971351 EFA06477.1 GEZM01057544 JAV72263.1 NEVH01016313 PNF25715.1 KB632367 ERL93662.1 GEBQ01028256 JAT11721.1 APGK01045785 APGK01045786 APGK01045787 KB741039 ENN74521.1 DS235200 EEB13187.1 LJIJ01000919 ODM93765.1 GFTR01008716 JAW07710.1 LNIX01000001 OXA64251.1 GDHC01014543 JAQ04086.1 CVRI01000014 CRK89749.1 GDAI01002259 JAI15344.1 JXUM01071164 JXUM01071165 KQ562647 KXJ75414.1 GDUN01001116 JAN94803.1 AAAB01008964 APCN01005201 CH477570 EAT38881.1 GDIP01206057 JAJ17345.1 GBXI01012528 JAD01764.1 DS231817 EDS39640.1 GBXI01010630 JAD03662.1 GDIP01061933 JAM41782.1 GDIP01237460 GDIQ01177437 JAI85941.1 JAK74288.1 GAMC01020547 JAB86008.1 GDIQ01239624 JAK12101.1 GDIP01114824 JAL88890.1 GDHF01029605 JAI22709.1 GDIQ01177438 JAK74287.1 GDIP01162064 JAJ61338.1 GDIQ01025902 JAN68835.1 GDIP01032786 JAM70929.1 GDIQ01220285 JAK31440.1 GDIQ01151756 JAK99969.1 GDIQ01050647 JAN44090.1 GDIQ01138761 JAL12965.1 GDIQ01144263 JAL07463.1 GDIQ01080184 JAN14553.1 GDIQ01132667 JAL19059.1 GDIP01149621 JAJ73781.1 GDIQ01157051 JAK94674.1 GDIP01203911 JAJ19491.1 GDIQ01253128 JAJ98596.1 LRGB01003341 KZS03126.1 GDIQ01145604 JAL06122.1 GDHF01027154 JAI25160.1 GDHF01015690 JAI36624.1 GAKP01009656 JAC49296.1 AXCM01007603 GDIQ01112112 JAL39614.1 GDIP01089529 JAM14186.1 CH902620 EDV32030.1 CH916372 EDV98773.1 ADMH02001723 ETN61299.1 CH940654 EDW57676.1 KRF77886.1 AXCN02000707 ATLV01021262 ATLV01021263 KE525315 KFB46207.1
KQ459053 KPJ04215.1 KQ461181 KPJ07756.1 RSAL01000042 RVE50792.1 GEZM01057545 JAV72261.1 KQ971351 EFA06477.1 GEZM01057544 JAV72263.1 NEVH01016313 PNF25715.1 KB632367 ERL93662.1 GEBQ01028256 JAT11721.1 APGK01045785 APGK01045786 APGK01045787 KB741039 ENN74521.1 DS235200 EEB13187.1 LJIJ01000919 ODM93765.1 GFTR01008716 JAW07710.1 LNIX01000001 OXA64251.1 GDHC01014543 JAQ04086.1 CVRI01000014 CRK89749.1 GDAI01002259 JAI15344.1 JXUM01071164 JXUM01071165 KQ562647 KXJ75414.1 GDUN01001116 JAN94803.1 AAAB01008964 APCN01005201 CH477570 EAT38881.1 GDIP01206057 JAJ17345.1 GBXI01012528 JAD01764.1 DS231817 EDS39640.1 GBXI01010630 JAD03662.1 GDIP01061933 JAM41782.1 GDIP01237460 GDIQ01177437 JAI85941.1 JAK74288.1 GAMC01020547 JAB86008.1 GDIQ01239624 JAK12101.1 GDIP01114824 JAL88890.1 GDHF01029605 JAI22709.1 GDIQ01177438 JAK74287.1 GDIP01162064 JAJ61338.1 GDIQ01025902 JAN68835.1 GDIP01032786 JAM70929.1 GDIQ01220285 JAK31440.1 GDIQ01151756 JAK99969.1 GDIQ01050647 JAN44090.1 GDIQ01138761 JAL12965.1 GDIQ01144263 JAL07463.1 GDIQ01080184 JAN14553.1 GDIQ01132667 JAL19059.1 GDIP01149621 JAJ73781.1 GDIQ01157051 JAK94674.1 GDIP01203911 JAJ19491.1 GDIQ01253128 JAJ98596.1 LRGB01003341 KZS03126.1 GDIQ01145604 JAL06122.1 GDHF01027154 JAI25160.1 GDHF01015690 JAI36624.1 GAKP01009656 JAC49296.1 AXCM01007603 GDIQ01112112 JAL39614.1 GDIP01089529 JAM14186.1 CH902620 EDV32030.1 CH916372 EDV98773.1 ADMH02001723 ETN61299.1 CH940654 EDW57676.1 KRF77886.1 AXCN02000707 ATLV01021262 ATLV01021263 KE525315 KFB46207.1
Proteomes
UP000218220
UP000007151
UP000053268
UP000053240
UP000283053
UP000192223
+ More
UP000007266 UP000235965 UP000030742 UP000019118 UP000009046 UP000094527 UP000198287 UP000183832 UP000069940 UP000249989 UP000075840 UP000075882 UP000076407 UP000008820 UP000075881 UP000002320 UP000076858 UP000075920 UP000069272 UP000075900 UP000075883 UP000076408 UP000075885 UP000092445 UP000007801 UP000075880 UP000001070 UP000000673 UP000008792 UP000075886 UP000030765
UP000007266 UP000235965 UP000030742 UP000019118 UP000009046 UP000094527 UP000198287 UP000183832 UP000069940 UP000249989 UP000075840 UP000075882 UP000076407 UP000008820 UP000075881 UP000002320 UP000076858 UP000075920 UP000069272 UP000075900 UP000075883 UP000076408 UP000075885 UP000092445 UP000007801 UP000075880 UP000001070 UP000000673 UP000008792 UP000075886 UP000030765
Interpro
IPR027417
P-loop_NTPase
+ More
IPR001680 WD40_repeat
IPR019775 WD40_repeat_CS
IPR017986 WD40_repeat_dom
IPR011047 Quinoprotein_ADH-like_supfam
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR007111 NACHT_NTPase
IPR036322 WD40_repeat_dom_sf
IPR000318 Nase_comp1_CS
IPR030934 Intein_C
IPR003593 AAA+_ATPase
IPR000436 Sushi_SCR_CCP_dom
IPR008197 WAP_dom
IPR035976 Sushi/SCR/CCP_sf
IPR001680 WD40_repeat
IPR019775 WD40_repeat_CS
IPR017986 WD40_repeat_dom
IPR011047 Quinoprotein_ADH-like_supfam
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR007111 NACHT_NTPase
IPR036322 WD40_repeat_dom_sf
IPR000318 Nase_comp1_CS
IPR030934 Intein_C
IPR003593 AAA+_ATPase
IPR000436 Sushi_SCR_CCP_dom
IPR008197 WAP_dom
IPR035976 Sushi/SCR/CCP_sf
Gene 3D
CDD
ProteinModelPortal
A0A2H1V5G8
A0A2A4JAS8
A0A212FLL4
A0A194QH02
A0A194QW14
A0A3S2NGX7
+ More
A0A1W4WH42 A0A1W4W6Q0 A0A1W4WID3 A0A1Y1LH95 D6WR21 A0A1Y1LF63 A0A2J7QAW1 U4UKL9 A0A1B6KJV3 N6U1N5 E0VII1 A0A1D2MLL7 A0A224XGX4 A0A226F4Y7 A0A146LA42 A0A1J1HNX5 A0A0K8TLW2 A0A182GL42 A0A1S4FLV2 A0A0P6ISD2 A0A1S4GW40 A0A182IED3 A0A182L2N3 A0A182X1F3 Q16WD7 A0A182K0E6 A0A0N7ZWD5 A0A0A1WRS3 B0W0A1 A0A0A1WXR7 A0A0P5Z4E0 A0A0P5L1U9 W8ANI1 A0A0P5GPH2 A0A0P5UFZ3 A0A0K8U7S4 A0A0P5LI65 A0A0P5D2C0 A0A0P6H1F6 A0A0P6B7Z9 A0A0P5IK15 A0A0P5NAM8 A0A0P6FFP5 A0A0P5NRK6 A0A0P5PUA0 A0A0P6DBI9 A0A0N8BYE9 A0A0P5DWT5 A0A0N8BPQ1 A0A0P5A522 A0A0P5FRE4 A0A164KBN3 A0A0P5N8V9 A0A0K8UFP0 A0A0K8VCE5 A0A182W7M3 A0A182F1B4 A0A034W167 A0A182RU62 A0A182MPC5 A0A0P5R2V8 A0A0P5WAA1 A0A182XZQ7 A0A182PJR6 A0A1A9Z411 B3MN52 A0A182IS92 B4JPE3 W5JEP8 B4M900 A0A182QHQ1 A0A084W7L3
A0A1W4WH42 A0A1W4W6Q0 A0A1W4WID3 A0A1Y1LH95 D6WR21 A0A1Y1LF63 A0A2J7QAW1 U4UKL9 A0A1B6KJV3 N6U1N5 E0VII1 A0A1D2MLL7 A0A224XGX4 A0A226F4Y7 A0A146LA42 A0A1J1HNX5 A0A0K8TLW2 A0A182GL42 A0A1S4FLV2 A0A0P6ISD2 A0A1S4GW40 A0A182IED3 A0A182L2N3 A0A182X1F3 Q16WD7 A0A182K0E6 A0A0N7ZWD5 A0A0A1WRS3 B0W0A1 A0A0A1WXR7 A0A0P5Z4E0 A0A0P5L1U9 W8ANI1 A0A0P5GPH2 A0A0P5UFZ3 A0A0K8U7S4 A0A0P5LI65 A0A0P5D2C0 A0A0P6H1F6 A0A0P6B7Z9 A0A0P5IK15 A0A0P5NAM8 A0A0P6FFP5 A0A0P5NRK6 A0A0P5PUA0 A0A0P6DBI9 A0A0N8BYE9 A0A0P5DWT5 A0A0N8BPQ1 A0A0P5A522 A0A0P5FRE4 A0A164KBN3 A0A0P5N8V9 A0A0K8UFP0 A0A0K8VCE5 A0A182W7M3 A0A182F1B4 A0A034W167 A0A182RU62 A0A182MPC5 A0A0P5R2V8 A0A0P5WAA1 A0A182XZQ7 A0A182PJR6 A0A1A9Z411 B3MN52 A0A182IS92 B4JPE3 W5JEP8 B4M900 A0A182QHQ1 A0A084W7L3
Ontologies
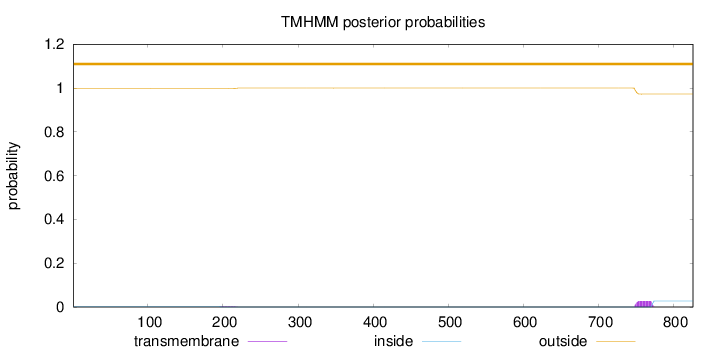
Topology
Length:
826
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.64524
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00183
outside
1 - 826
Population Genetic Test Statistics
Pi
238.824888
Theta
151.401275
Tajima's D
0.206911
CLR
1.043614
CSRT
0.425828708564572
Interpretation
Uncertain
