Gene
KWMTBOMO12188
Pre Gene Modal
BGIBMGA004230
Annotation
hypothetical_protein_RR46_07967_[Papilio_xuthus]
Location in the cell
Extracellular Reliability : 2.628
Sequence
CDS
ATGATGTTCCACGTGAAAGTTTTTTTTGCTTGTCACATACTATTTGGCATATACATGGTTCTGAGTACTAAGCCAGAGCCGACACCTGTAACATTCACATTCAAAGAATATTTATATAAAGATTCACCAAAAAATGAAATTACTTACAAGGAATTCGAGACCGCATGTGAACAAAGCAGCGCCTGTAGTTACCAAAAAGGTTTGCAAAAAACTCGATGCGTCCGAGAGTGTATTTCTCCATCGTGCTACAAAGAAATCTATCAAGCAGATCCGCTTGAAGAAGGTGAAATCGACGTCCGATTGAACTCGTTCAAGGGTTGTTTTGCACAGAGAGCGGGTCGTTCGAGGAATTGA
Protein
MMFHVKVFFACHILFGIYMVLSTKPEPTPVTFTFKEYLYKDSPKNEITYKEFETACEQSSACSYQKGLQKTRCVRECISPSCYKEIYQADPLEEGEIDVRLNSFKGCFAQRAGRSRN
Summary
Uniprot
H9J3Z1
A0A194QF88
A0A194QRZ0
A0A2H1VGE4
A0A212EYY5
A0A182FHK5
+ More
A0A067R209 A0A1S4F9T2 A0A2J7R2X0 A0A1B6GNQ5 A0A1B6HDR1 E2C392 B0W3Q3 A0A0C9QBT7 Q17AU5 A0A1B6MJ43 A0A1B0D147 A0A0L7RJB7 A0A182KPZ3 A0A1W7R751 A0A182VEQ4 A0A182MUE2 A0A0N0U3E3 A0A182K910 A0A182RZ06 A0A151X0F1 A0A182I272 A0A182XMD9 A0A182WLA2 A0A146KP62 A0A195CQ87 A0A195EMP4 A0A195BWJ7 Q7Q6H7 A0A0A9YQP2 E9IM74 E2A2N3 A0A195F9W1 A0A182NJF3 F4WX38 A0A182YJ87 A0A158NCI5 A0A3L8DGH9 A0A182QHC5 A0A0K8SY26 A0A2A3EPI3 A0A088AVR9 W5J491 T1IFM2 A0A1A9WLD8 A0A0T6BHM6 A0A2K8JMF2 A0A1B0GF63 A0A1I8NLE8 A0A1B0AJB5 D6WLF6 A0A1A9US10 A0A1I8NH20 A0A0K8W7M9 A0A0L0BQL3 E0VZ76 A0A034WVM7 A0A1Y1LI61 B5DZL5 B4H716 A0A0P4W9G5 B4MI08 A0A1W4V438 B4PKI2 B3P0G7 B4QV47 B3LXE9 B4IBD7 Q9VEG3 B5RJ07 A0A336K8I9 B4JUG1 A0A3B0K2D3 N6TNM9 B4M5G9 B4KDQ3 A0A0P5R869 A0A0N7ZGQ7 A0A0M4EQP1 A0A0Q9W2J6 A0A1D2MZF7 A0A0P5J1J8 A0A210PRK5 A0A369S5R6 A0A0C9QYD2 B3S508 A0A0B7B0G0 A0A310SXH7
A0A067R209 A0A1S4F9T2 A0A2J7R2X0 A0A1B6GNQ5 A0A1B6HDR1 E2C392 B0W3Q3 A0A0C9QBT7 Q17AU5 A0A1B6MJ43 A0A1B0D147 A0A0L7RJB7 A0A182KPZ3 A0A1W7R751 A0A182VEQ4 A0A182MUE2 A0A0N0U3E3 A0A182K910 A0A182RZ06 A0A151X0F1 A0A182I272 A0A182XMD9 A0A182WLA2 A0A146KP62 A0A195CQ87 A0A195EMP4 A0A195BWJ7 Q7Q6H7 A0A0A9YQP2 E9IM74 E2A2N3 A0A195F9W1 A0A182NJF3 F4WX38 A0A182YJ87 A0A158NCI5 A0A3L8DGH9 A0A182QHC5 A0A0K8SY26 A0A2A3EPI3 A0A088AVR9 W5J491 T1IFM2 A0A1A9WLD8 A0A0T6BHM6 A0A2K8JMF2 A0A1B0GF63 A0A1I8NLE8 A0A1B0AJB5 D6WLF6 A0A1A9US10 A0A1I8NH20 A0A0K8W7M9 A0A0L0BQL3 E0VZ76 A0A034WVM7 A0A1Y1LI61 B5DZL5 B4H716 A0A0P4W9G5 B4MI08 A0A1W4V438 B4PKI2 B3P0G7 B4QV47 B3LXE9 B4IBD7 Q9VEG3 B5RJ07 A0A336K8I9 B4JUG1 A0A3B0K2D3 N6TNM9 B4M5G9 B4KDQ3 A0A0P5R869 A0A0N7ZGQ7 A0A0M4EQP1 A0A0Q9W2J6 A0A1D2MZF7 A0A0P5J1J8 A0A210PRK5 A0A369S5R6 A0A0C9QYD2 B3S508 A0A0B7B0G0 A0A310SXH7
Pubmed
19121390
26354079
22118469
24845553
20798317
17510324
+ More
20966253 26823975 12364791 14747013 17210077 25401762 21282665 21719571 25244985 21347285 30249741 20920257 23761445 18362917 19820115 25315136 26108605 20566863 25348373 28004739 15632085 17994087 18057021 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 23537049 27289101 28812685 30042472 18719581
20966253 26823975 12364791 14747013 17210077 25401762 21282665 21719571 25244985 21347285 30249741 20920257 23761445 18362917 19820115 25315136 26108605 20566863 25348373 28004739 15632085 17994087 18057021 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 23537049 27289101 28812685 30042472 18719581
EMBL
BABH01040254
BABH01040255
KQ459053
KPJ04208.1
KQ461181
KPJ07750.1
+ More
ODYU01002435 SOQ39923.1 AGBW02011414 OWR46700.1 KK853080 KDR11698.1 NEVH01007824 PNF35182.1 GECZ01005729 JAS64040.1 GECU01034869 GECU01028565 GECU01015983 JAS72837.1 JAS79141.1 JAS91723.1 GL452292 EFN77624.1 DS231833 EDS32200.1 GBYB01000704 JAG70471.1 CH477330 EAT43363.1 GEBQ01004000 JAT35977.1 AJVK01010196 AJVK01010197 AJVK01010198 KQ414582 KOC70878.1 GEHC01000685 JAV46960.1 AXCM01003810 KQ435922 KOX68451.1 KQ982614 KYQ53836.1 APCN01000126 GDHC01021567 JAP97061.1 KQ977408 KYN02913.1 KQ978691 KYN29172.1 KQ976396 KYM92957.1 AAAB01008960 EAA11044.3 GBHO01009096 GBHO01009095 JAG34508.1 JAG34509.1 GL764129 EFZ18371.1 GL436181 EFN72298.1 KQ981727 KYN37002.1 GL888417 EGI61281.1 ADTU01011908 ADTU01011909 ADTU01011910 QOIP01000008 RLU19421.1 AXCN02000446 GBRD01008006 JAG57815.1 KZ288198 PBC33675.1 ADMH02002096 ETN59242.1 ACPB03005594 LJIG01000115 KRT86825.1 KY031238 ATU82989.1 CCAG010012764 KQ971343 EFA03457.1 GDHF01005121 JAI47193.1 JRES01001517 KNC22301.1 DS235850 EEB18682.1 GAKP01000707 JAC58245.1 GEZM01054922 JAV73339.1 CM000071 EDY69595.1 CH479216 EDW33652.1 GDRN01075918 JAI62973.1 CH961255 CH964251 EDW71747.1 EDW83076.1 CM000160 EDW95821.1 CH954181 EDV48682.1 CM000364 EDX12530.1 CH902617 EDV42793.2 CH480827 EDW44695.1 AE014297 AAF55461.1 BT044281 ACH92346.1 UFQS01000086 UFQT01000086 SSW99073.1 SSX19455.1 CH916374 EDV91131.1 OUUW01000014 SPP88455.1 APGK01028397 KB740686 KB631879 ENN79578.1 ERL86902.1 CH940652 EDW59880.1 CH933806 EDW13887.1 GDIQ01111310 JAL40416.1 GDIP01247081 JAI76320.1 CP012526 ALC46685.1 KRF79138.1 LJIJ01000361 ODM98380.1 GDIQ01204425 JAK47300.1 NEDP02005542 OWF39120.1 NOWV01000058 RDD42257.1 GBYB01000705 JAG70472.1 DS985250 EDV22183.1 HACG01038931 CEK85796.1 KQ759817 OAD62786.1
ODYU01002435 SOQ39923.1 AGBW02011414 OWR46700.1 KK853080 KDR11698.1 NEVH01007824 PNF35182.1 GECZ01005729 JAS64040.1 GECU01034869 GECU01028565 GECU01015983 JAS72837.1 JAS79141.1 JAS91723.1 GL452292 EFN77624.1 DS231833 EDS32200.1 GBYB01000704 JAG70471.1 CH477330 EAT43363.1 GEBQ01004000 JAT35977.1 AJVK01010196 AJVK01010197 AJVK01010198 KQ414582 KOC70878.1 GEHC01000685 JAV46960.1 AXCM01003810 KQ435922 KOX68451.1 KQ982614 KYQ53836.1 APCN01000126 GDHC01021567 JAP97061.1 KQ977408 KYN02913.1 KQ978691 KYN29172.1 KQ976396 KYM92957.1 AAAB01008960 EAA11044.3 GBHO01009096 GBHO01009095 JAG34508.1 JAG34509.1 GL764129 EFZ18371.1 GL436181 EFN72298.1 KQ981727 KYN37002.1 GL888417 EGI61281.1 ADTU01011908 ADTU01011909 ADTU01011910 QOIP01000008 RLU19421.1 AXCN02000446 GBRD01008006 JAG57815.1 KZ288198 PBC33675.1 ADMH02002096 ETN59242.1 ACPB03005594 LJIG01000115 KRT86825.1 KY031238 ATU82989.1 CCAG010012764 KQ971343 EFA03457.1 GDHF01005121 JAI47193.1 JRES01001517 KNC22301.1 DS235850 EEB18682.1 GAKP01000707 JAC58245.1 GEZM01054922 JAV73339.1 CM000071 EDY69595.1 CH479216 EDW33652.1 GDRN01075918 JAI62973.1 CH961255 CH964251 EDW71747.1 EDW83076.1 CM000160 EDW95821.1 CH954181 EDV48682.1 CM000364 EDX12530.1 CH902617 EDV42793.2 CH480827 EDW44695.1 AE014297 AAF55461.1 BT044281 ACH92346.1 UFQS01000086 UFQT01000086 SSW99073.1 SSX19455.1 CH916374 EDV91131.1 OUUW01000014 SPP88455.1 APGK01028397 KB740686 KB631879 ENN79578.1 ERL86902.1 CH940652 EDW59880.1 CH933806 EDW13887.1 GDIQ01111310 JAL40416.1 GDIP01247081 JAI76320.1 CP012526 ALC46685.1 KRF79138.1 LJIJ01000361 ODM98380.1 GDIQ01204425 JAK47300.1 NEDP02005542 OWF39120.1 NOWV01000058 RDD42257.1 GBYB01000705 JAG70472.1 DS985250 EDV22183.1 HACG01038931 CEK85796.1 KQ759817 OAD62786.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000007151
UP000069272
UP000027135
+ More
UP000235965 UP000008237 UP000002320 UP000008820 UP000092462 UP000053825 UP000075882 UP000075903 UP000075883 UP000053105 UP000075881 UP000075900 UP000075809 UP000075840 UP000076407 UP000075920 UP000078542 UP000078492 UP000078540 UP000007062 UP000000311 UP000078541 UP000075884 UP000007755 UP000076408 UP000005205 UP000279307 UP000075886 UP000242457 UP000005203 UP000000673 UP000015103 UP000091820 UP000092444 UP000095300 UP000092445 UP000007266 UP000078200 UP000095301 UP000037069 UP000009046 UP000001819 UP000008744 UP000007798 UP000192221 UP000002282 UP000008711 UP000000304 UP000007801 UP000001292 UP000000803 UP000001070 UP000268350 UP000019118 UP000030742 UP000008792 UP000009192 UP000092553 UP000094527 UP000242188 UP000253843 UP000009022
UP000235965 UP000008237 UP000002320 UP000008820 UP000092462 UP000053825 UP000075882 UP000075903 UP000075883 UP000053105 UP000075881 UP000075900 UP000075809 UP000075840 UP000076407 UP000075920 UP000078542 UP000078492 UP000078540 UP000007062 UP000000311 UP000078541 UP000075884 UP000007755 UP000076408 UP000005205 UP000279307 UP000075886 UP000242457 UP000005203 UP000000673 UP000015103 UP000091820 UP000092444 UP000095300 UP000092445 UP000007266 UP000078200 UP000095301 UP000037069 UP000009046 UP000001819 UP000008744 UP000007798 UP000192221 UP000002282 UP000008711 UP000000304 UP000007801 UP000001292 UP000000803 UP000001070 UP000268350 UP000019118 UP000030742 UP000008792 UP000009192 UP000092553 UP000094527 UP000242188 UP000253843 UP000009022
Interpro
SUPFAM
SSF48371
SSF48371
Gene 3D
ProteinModelPortal
H9J3Z1
A0A194QF88
A0A194QRZ0
A0A2H1VGE4
A0A212EYY5
A0A182FHK5
+ More
A0A067R209 A0A1S4F9T2 A0A2J7R2X0 A0A1B6GNQ5 A0A1B6HDR1 E2C392 B0W3Q3 A0A0C9QBT7 Q17AU5 A0A1B6MJ43 A0A1B0D147 A0A0L7RJB7 A0A182KPZ3 A0A1W7R751 A0A182VEQ4 A0A182MUE2 A0A0N0U3E3 A0A182K910 A0A182RZ06 A0A151X0F1 A0A182I272 A0A182XMD9 A0A182WLA2 A0A146KP62 A0A195CQ87 A0A195EMP4 A0A195BWJ7 Q7Q6H7 A0A0A9YQP2 E9IM74 E2A2N3 A0A195F9W1 A0A182NJF3 F4WX38 A0A182YJ87 A0A158NCI5 A0A3L8DGH9 A0A182QHC5 A0A0K8SY26 A0A2A3EPI3 A0A088AVR9 W5J491 T1IFM2 A0A1A9WLD8 A0A0T6BHM6 A0A2K8JMF2 A0A1B0GF63 A0A1I8NLE8 A0A1B0AJB5 D6WLF6 A0A1A9US10 A0A1I8NH20 A0A0K8W7M9 A0A0L0BQL3 E0VZ76 A0A034WVM7 A0A1Y1LI61 B5DZL5 B4H716 A0A0P4W9G5 B4MI08 A0A1W4V438 B4PKI2 B3P0G7 B4QV47 B3LXE9 B4IBD7 Q9VEG3 B5RJ07 A0A336K8I9 B4JUG1 A0A3B0K2D3 N6TNM9 B4M5G9 B4KDQ3 A0A0P5R869 A0A0N7ZGQ7 A0A0M4EQP1 A0A0Q9W2J6 A0A1D2MZF7 A0A0P5J1J8 A0A210PRK5 A0A369S5R6 A0A0C9QYD2 B3S508 A0A0B7B0G0 A0A310SXH7
A0A067R209 A0A1S4F9T2 A0A2J7R2X0 A0A1B6GNQ5 A0A1B6HDR1 E2C392 B0W3Q3 A0A0C9QBT7 Q17AU5 A0A1B6MJ43 A0A1B0D147 A0A0L7RJB7 A0A182KPZ3 A0A1W7R751 A0A182VEQ4 A0A182MUE2 A0A0N0U3E3 A0A182K910 A0A182RZ06 A0A151X0F1 A0A182I272 A0A182XMD9 A0A182WLA2 A0A146KP62 A0A195CQ87 A0A195EMP4 A0A195BWJ7 Q7Q6H7 A0A0A9YQP2 E9IM74 E2A2N3 A0A195F9W1 A0A182NJF3 F4WX38 A0A182YJ87 A0A158NCI5 A0A3L8DGH9 A0A182QHC5 A0A0K8SY26 A0A2A3EPI3 A0A088AVR9 W5J491 T1IFM2 A0A1A9WLD8 A0A0T6BHM6 A0A2K8JMF2 A0A1B0GF63 A0A1I8NLE8 A0A1B0AJB5 D6WLF6 A0A1A9US10 A0A1I8NH20 A0A0K8W7M9 A0A0L0BQL3 E0VZ76 A0A034WVM7 A0A1Y1LI61 B5DZL5 B4H716 A0A0P4W9G5 B4MI08 A0A1W4V438 B4PKI2 B3P0G7 B4QV47 B3LXE9 B4IBD7 Q9VEG3 B5RJ07 A0A336K8I9 B4JUG1 A0A3B0K2D3 N6TNM9 B4M5G9 B4KDQ3 A0A0P5R869 A0A0N7ZGQ7 A0A0M4EQP1 A0A0Q9W2J6 A0A1D2MZF7 A0A0P5J1J8 A0A210PRK5 A0A369S5R6 A0A0C9QYD2 B3S508 A0A0B7B0G0 A0A310SXH7
Ontologies
Topology
SignalP
Position: 1 - 22,
Likelihood: 0.899247
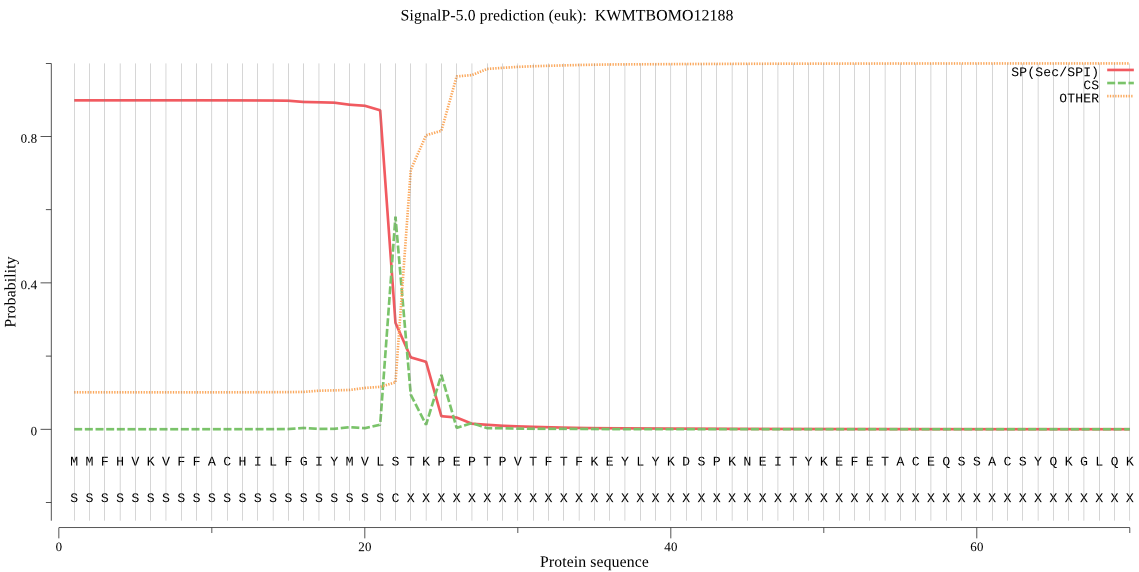
Length:
117
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
5.34661
Exp number, first 60 AAs:
5.34661
Total prob of N-in:
0.34182
outside
1 - 117

Population Genetic Test Statistics
Pi
176.18558
Theta
179.3362
Tajima's D
-0.576183
CLR
0.230405
CSRT
0.223238838058097
Interpretation
Uncertain
