Gene
KWMTBOMO12187
Pre Gene Modal
BGIBMGA004215
Annotation
PREDICTED:_uncharacterized_protein_LOC101735402_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 3.486
Sequence
CDS
ATGCGTTCGTTGTTGGCTTACTACGCGGACGCGGTGATGTCGGTGGCGGCTAAGGGCGCCGGCTGCGCGCTCAGCTGCAGCGGCAGGGGCGACTGCATGAACGGCACCTGTCTCTGCGAGATCCGGTACTCGGGCGAGGGTTGCGCCGGACCTAACCTGCCGTATCATGCGTGTATTGGCGGTGTTTTCCTCGTCGTCGCCTTCGTGTGCGCCGTCCAACTGATGATATGCGTTGTCACGGAGTACAGGAAACTAAAGGCTCCGACTTTCCTGAGGGCTTGCAAGGTCACGACCCAGAAGATGCTGTATCTGGTCGCTTTCCTCGCCTCCCTGATACGGGGAGCTTACTTCGTTTCGCCGTCTGCTTTTCAAGAGGGATGGGCGACCAGTCTTCTATCGGCTTATTATCCCCTGTTGATGTCTGGATCGTCGCTGATCGTGTGCTTTTGGGCCGAGGTCTCAAGGTAA
Protein
MRSLLAYYADAVMSVAAKGAGCALSCSGRGDCMNGTCLCEIRYSGEGCAGPNLPYHACIGGVFLVVAFVCAVQLMICVVTEYRKLKAPTFLRACKVTTQKMLYLVAFLASLIRGAYFVSPSAFQEGWATSLLSAYYPLLMSGSSLIVCFWAEVSR
Summary
Uniprot
H9J3X6
A0A2H1VS50
A0A194QFE6
A0A194QQX4
A0A3S2NLL7
A0A2A4IWD1
+ More
A0A1E1VY05 A0A2W1BHR7 A0A1Y1K655 D6WSX5 A0A151INS8 A0A154PL39 A0A026WJH2 E1ZYP8 A0A2A3EHR1 V9IJG0 A0A087ZQA5 E2AXV2 A0A2P8Z580 A0A151WPN3 A0A0C9QPM1 A0A182IQP9 A0A2J7QQT6 A0A310S771 A0A0N0BHI5 A0A067QZ96 A0A2J7QQS1 B0WNA1 A0A0K8TT33 A0A023ETJ5 A0A182SNI2 U5ENJ3 E2BS63 Q17CM2 A0A1S4F7W4 A0A1Q3FYM1 A0A1Q3FYL7 A0A1B0CSS5 A0A1L8DSD8 A0A2M4A5A6 A0A1I8PW21 A0A2M4BKS0 A0A2M3Z2D3 W5JGN0 A0A182F0P4 A0A2M4BKG2 T1PDL3 A0A182WH73 A0A182PFS3 A0A1S4H150 A0A182VDQ5 A0A182KUF5 A0A182I571 Q7PVU7 A0A182UJ00 A0A182XKX3 A0A182NF55 A0A182Y9T1 A0A182MHL2 A0A084VMX7 A0A182QFZ3 A0A1B6MIT4 A0A0K8U458 A0A1A9UH41 A0A1A9XT66 A0A1A9ZFA8 A0A1B0BN40 A0A0L0BMK7 A0A1A9W5K5 A0A0V0G7S9 A0A224XAM9 A0A1B6J7N9 F4WYE3 A0A1B6JM06 A0A1B6HC48 A0A195EUR8 N6T2J8 A0A195ECR6 A0A195B0A8 A0A158NXP7 A0A0A9YKT4 W8B721 A0A0A1XNN5 A0A0A1XN77 E0VII0 B4M9H8 A0A0K8UK01 A0A3B0JJ67 B4GSB9 Q29KF6 A0A0M4EBL4 B4I2S1 B4Q8M2 Q9VQI2 B4NXA0 B3NA79 B4N000 B4KH11 A0A1B6C8I6 A0A1B6GD71
A0A1E1VY05 A0A2W1BHR7 A0A1Y1K655 D6WSX5 A0A151INS8 A0A154PL39 A0A026WJH2 E1ZYP8 A0A2A3EHR1 V9IJG0 A0A087ZQA5 E2AXV2 A0A2P8Z580 A0A151WPN3 A0A0C9QPM1 A0A182IQP9 A0A2J7QQT6 A0A310S771 A0A0N0BHI5 A0A067QZ96 A0A2J7QQS1 B0WNA1 A0A0K8TT33 A0A023ETJ5 A0A182SNI2 U5ENJ3 E2BS63 Q17CM2 A0A1S4F7W4 A0A1Q3FYM1 A0A1Q3FYL7 A0A1B0CSS5 A0A1L8DSD8 A0A2M4A5A6 A0A1I8PW21 A0A2M4BKS0 A0A2M3Z2D3 W5JGN0 A0A182F0P4 A0A2M4BKG2 T1PDL3 A0A182WH73 A0A182PFS3 A0A1S4H150 A0A182VDQ5 A0A182KUF5 A0A182I571 Q7PVU7 A0A182UJ00 A0A182XKX3 A0A182NF55 A0A182Y9T1 A0A182MHL2 A0A084VMX7 A0A182QFZ3 A0A1B6MIT4 A0A0K8U458 A0A1A9UH41 A0A1A9XT66 A0A1A9ZFA8 A0A1B0BN40 A0A0L0BMK7 A0A1A9W5K5 A0A0V0G7S9 A0A224XAM9 A0A1B6J7N9 F4WYE3 A0A1B6JM06 A0A1B6HC48 A0A195EUR8 N6T2J8 A0A195ECR6 A0A195B0A8 A0A158NXP7 A0A0A9YKT4 W8B721 A0A0A1XNN5 A0A0A1XN77 E0VII0 B4M9H8 A0A0K8UK01 A0A3B0JJ67 B4GSB9 Q29KF6 A0A0M4EBL4 B4I2S1 B4Q8M2 Q9VQI2 B4NXA0 B3NA79 B4N000 B4KH11 A0A1B6C8I6 A0A1B6GD71
Pubmed
19121390
26354079
28756777
28004739
18362917
19820115
+ More
24508170 30249741 20798317 29403074 24845553 26369729 24945155 17510324 20920257 23761445 25315136 12364791 20966253 25244985 24438588 26108605 21719571 23537049 21347285 25401762 26823975 24495485 25830018 20566863 17994087 18057021 15632085 23185243 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304
24508170 30249741 20798317 29403074 24845553 26369729 24945155 17510324 20920257 23761445 25315136 12364791 20966253 25244985 24438588 26108605 21719571 23537049 21347285 25401762 26823975 24495485 25830018 20566863 17994087 18057021 15632085 23185243 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304
EMBL
BABH01040248
ODYU01004132
SOQ43683.1
KQ459053
KPJ04207.1
KQ461181
+ More
KPJ07749.1 RSAL01000042 RVE50782.1 NWSH01006460 PCG63413.1 GDQN01011454 JAT79600.1 KZ150117 PZC73255.1 GEZM01097310 JAV54277.1 KQ971352 EFA06336.1 KQ976914 KYN07095.1 KQ434939 KZC12184.1 KK107168 QOIP01000001 EZA56197.1 RLU26724.1 GL435242 EFN73588.1 KZ288254 PBC30726.1 JR050389 AEY61263.1 GL443736 EFN61716.1 PYGN01000190 PSN51653.1 KQ982851 KYQ49862.1 GBYB01005569 JAG75336.1 AXCP01007770 NEVH01012083 PNF30927.1 KQ766333 OAD53704.1 KQ435750 KOX76311.1 KK853083 KDR11659.1 PNF30922.1 DS232009 EDS31550.1 GDAI01000518 JAI17085.1 GAPW01000933 JAC12665.1 GANO01004053 JAB55818.1 GL450151 EFN81455.1 CH477307 EAT44075.1 GFDL01002398 JAV32647.1 GFDL01002404 JAV32641.1 AJWK01026545 GFDF01004725 JAV09359.1 GGFK01002638 MBW35959.1 GGFJ01004410 MBW53551.1 GGFM01001847 MBW22598.1 ADMH02001466 ETN62478.1 GGFJ01004409 MBW53550.1 KA646225 AFP60854.1 AAAB01008984 APCN01003794 APCN01003795 EAA14825.4 AXCM01007284 ATLV01014632 KE524975 KFB39321.1 AXCN02000829 GEBQ01004154 JAT35823.1 GDHF01030815 JAI21499.1 JXJN01017179 JRES01001649 KNC21163.1 GECL01002072 JAP04052.1 GFTR01007031 JAW09395.1 GECU01012524 JAS95182.1 GL888440 EGI60795.1 GECU01007400 JAT00307.1 GECU01035453 JAS72253.1 KQ981958 KYN32005.1 APGK01054741 APGK01054742 APGK01054743 APGK01054744 APGK01054745 APGK01054746 APGK01054747 KB741253 ENN71778.1 KQ979074 KYN23015.1 KQ976692 KYM77725.1 ADTU01003275 ADTU01003276 GBHO01011363 GBRD01003907 GDHC01015330 JAG32241.1 JAG61914.1 JAQ03299.1 GAMC01013674 GAMC01013672 JAB92883.1 GBXI01001802 JAD12490.1 GBXI01013344 GBXI01003451 GBXI01001890 JAD00948.1 JAD10841.1 JAD12402.1 DS235200 EEB13186.1 CH940654 EDW57854.1 KRF77977.1 KRF77979.1 GDHF01026922 GDHF01025277 JAI25392.1 JAI27037.1 OUUW01000006 SPP82397.1 CH479189 EDW25666.1 CH379061 EAL33219.1 KRT04816.1 KRT04817.1 CP012523 ALC40001.1 CH480820 EDW54066.1 CM000361 CM002910 EDX03539.1 KMY87764.1 AE014134 BT006012 AAF51189.1 AAO74695.1 CM000157 EDW87457.1 KRJ97161.1 KRJ97162.1 CH954177 EDV57542.1 KQS70014.1 KQS70015.1 CH963920 EDW77935.1 CH933807 EDW12222.1 KRG03157.1 GEDC01027495 JAS09803.1 GECZ01022283 GECZ01009377 JAS47486.1 JAS60392.1
KPJ07749.1 RSAL01000042 RVE50782.1 NWSH01006460 PCG63413.1 GDQN01011454 JAT79600.1 KZ150117 PZC73255.1 GEZM01097310 JAV54277.1 KQ971352 EFA06336.1 KQ976914 KYN07095.1 KQ434939 KZC12184.1 KK107168 QOIP01000001 EZA56197.1 RLU26724.1 GL435242 EFN73588.1 KZ288254 PBC30726.1 JR050389 AEY61263.1 GL443736 EFN61716.1 PYGN01000190 PSN51653.1 KQ982851 KYQ49862.1 GBYB01005569 JAG75336.1 AXCP01007770 NEVH01012083 PNF30927.1 KQ766333 OAD53704.1 KQ435750 KOX76311.1 KK853083 KDR11659.1 PNF30922.1 DS232009 EDS31550.1 GDAI01000518 JAI17085.1 GAPW01000933 JAC12665.1 GANO01004053 JAB55818.1 GL450151 EFN81455.1 CH477307 EAT44075.1 GFDL01002398 JAV32647.1 GFDL01002404 JAV32641.1 AJWK01026545 GFDF01004725 JAV09359.1 GGFK01002638 MBW35959.1 GGFJ01004410 MBW53551.1 GGFM01001847 MBW22598.1 ADMH02001466 ETN62478.1 GGFJ01004409 MBW53550.1 KA646225 AFP60854.1 AAAB01008984 APCN01003794 APCN01003795 EAA14825.4 AXCM01007284 ATLV01014632 KE524975 KFB39321.1 AXCN02000829 GEBQ01004154 JAT35823.1 GDHF01030815 JAI21499.1 JXJN01017179 JRES01001649 KNC21163.1 GECL01002072 JAP04052.1 GFTR01007031 JAW09395.1 GECU01012524 JAS95182.1 GL888440 EGI60795.1 GECU01007400 JAT00307.1 GECU01035453 JAS72253.1 KQ981958 KYN32005.1 APGK01054741 APGK01054742 APGK01054743 APGK01054744 APGK01054745 APGK01054746 APGK01054747 KB741253 ENN71778.1 KQ979074 KYN23015.1 KQ976692 KYM77725.1 ADTU01003275 ADTU01003276 GBHO01011363 GBRD01003907 GDHC01015330 JAG32241.1 JAG61914.1 JAQ03299.1 GAMC01013674 GAMC01013672 JAB92883.1 GBXI01001802 JAD12490.1 GBXI01013344 GBXI01003451 GBXI01001890 JAD00948.1 JAD10841.1 JAD12402.1 DS235200 EEB13186.1 CH940654 EDW57854.1 KRF77977.1 KRF77979.1 GDHF01026922 GDHF01025277 JAI25392.1 JAI27037.1 OUUW01000006 SPP82397.1 CH479189 EDW25666.1 CH379061 EAL33219.1 KRT04816.1 KRT04817.1 CP012523 ALC40001.1 CH480820 EDW54066.1 CM000361 CM002910 EDX03539.1 KMY87764.1 AE014134 BT006012 AAF51189.1 AAO74695.1 CM000157 EDW87457.1 KRJ97161.1 KRJ97162.1 CH954177 EDV57542.1 KQS70014.1 KQS70015.1 CH963920 EDW77935.1 CH933807 EDW12222.1 KRG03157.1 GEDC01027495 JAS09803.1 GECZ01022283 GECZ01009377 JAS47486.1 JAS60392.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000283053
UP000218220
UP000007266
+ More
UP000078542 UP000076502 UP000053097 UP000279307 UP000000311 UP000242457 UP000005203 UP000245037 UP000075809 UP000075880 UP000235965 UP000053105 UP000027135 UP000002320 UP000075901 UP000008237 UP000008820 UP000092461 UP000095300 UP000000673 UP000069272 UP000095301 UP000075920 UP000075885 UP000075903 UP000075882 UP000075840 UP000007062 UP000075902 UP000076407 UP000075884 UP000076408 UP000075883 UP000030765 UP000075886 UP000078200 UP000092443 UP000092445 UP000092460 UP000037069 UP000091820 UP000007755 UP000078541 UP000019118 UP000078492 UP000078540 UP000005205 UP000009046 UP000008792 UP000268350 UP000008744 UP000001819 UP000092553 UP000001292 UP000000304 UP000000803 UP000002282 UP000008711 UP000007798 UP000009192
UP000078542 UP000076502 UP000053097 UP000279307 UP000000311 UP000242457 UP000005203 UP000245037 UP000075809 UP000075880 UP000235965 UP000053105 UP000027135 UP000002320 UP000075901 UP000008237 UP000008820 UP000092461 UP000095300 UP000000673 UP000069272 UP000095301 UP000075920 UP000075885 UP000075903 UP000075882 UP000075840 UP000007062 UP000075902 UP000076407 UP000075884 UP000076408 UP000075883 UP000030765 UP000075886 UP000078200 UP000092443 UP000092445 UP000092460 UP000037069 UP000091820 UP000007755 UP000078541 UP000019118 UP000078492 UP000078540 UP000005205 UP000009046 UP000008792 UP000268350 UP000008744 UP000001819 UP000092553 UP000001292 UP000000304 UP000000803 UP000002282 UP000008711 UP000007798 UP000009192
Interpro
Gene 3D
ProteinModelPortal
H9J3X6
A0A2H1VS50
A0A194QFE6
A0A194QQX4
A0A3S2NLL7
A0A2A4IWD1
+ More
A0A1E1VY05 A0A2W1BHR7 A0A1Y1K655 D6WSX5 A0A151INS8 A0A154PL39 A0A026WJH2 E1ZYP8 A0A2A3EHR1 V9IJG0 A0A087ZQA5 E2AXV2 A0A2P8Z580 A0A151WPN3 A0A0C9QPM1 A0A182IQP9 A0A2J7QQT6 A0A310S771 A0A0N0BHI5 A0A067QZ96 A0A2J7QQS1 B0WNA1 A0A0K8TT33 A0A023ETJ5 A0A182SNI2 U5ENJ3 E2BS63 Q17CM2 A0A1S4F7W4 A0A1Q3FYM1 A0A1Q3FYL7 A0A1B0CSS5 A0A1L8DSD8 A0A2M4A5A6 A0A1I8PW21 A0A2M4BKS0 A0A2M3Z2D3 W5JGN0 A0A182F0P4 A0A2M4BKG2 T1PDL3 A0A182WH73 A0A182PFS3 A0A1S4H150 A0A182VDQ5 A0A182KUF5 A0A182I571 Q7PVU7 A0A182UJ00 A0A182XKX3 A0A182NF55 A0A182Y9T1 A0A182MHL2 A0A084VMX7 A0A182QFZ3 A0A1B6MIT4 A0A0K8U458 A0A1A9UH41 A0A1A9XT66 A0A1A9ZFA8 A0A1B0BN40 A0A0L0BMK7 A0A1A9W5K5 A0A0V0G7S9 A0A224XAM9 A0A1B6J7N9 F4WYE3 A0A1B6JM06 A0A1B6HC48 A0A195EUR8 N6T2J8 A0A195ECR6 A0A195B0A8 A0A158NXP7 A0A0A9YKT4 W8B721 A0A0A1XNN5 A0A0A1XN77 E0VII0 B4M9H8 A0A0K8UK01 A0A3B0JJ67 B4GSB9 Q29KF6 A0A0M4EBL4 B4I2S1 B4Q8M2 Q9VQI2 B4NXA0 B3NA79 B4N000 B4KH11 A0A1B6C8I6 A0A1B6GD71
A0A1E1VY05 A0A2W1BHR7 A0A1Y1K655 D6WSX5 A0A151INS8 A0A154PL39 A0A026WJH2 E1ZYP8 A0A2A3EHR1 V9IJG0 A0A087ZQA5 E2AXV2 A0A2P8Z580 A0A151WPN3 A0A0C9QPM1 A0A182IQP9 A0A2J7QQT6 A0A310S771 A0A0N0BHI5 A0A067QZ96 A0A2J7QQS1 B0WNA1 A0A0K8TT33 A0A023ETJ5 A0A182SNI2 U5ENJ3 E2BS63 Q17CM2 A0A1S4F7W4 A0A1Q3FYM1 A0A1Q3FYL7 A0A1B0CSS5 A0A1L8DSD8 A0A2M4A5A6 A0A1I8PW21 A0A2M4BKS0 A0A2M3Z2D3 W5JGN0 A0A182F0P4 A0A2M4BKG2 T1PDL3 A0A182WH73 A0A182PFS3 A0A1S4H150 A0A182VDQ5 A0A182KUF5 A0A182I571 Q7PVU7 A0A182UJ00 A0A182XKX3 A0A182NF55 A0A182Y9T1 A0A182MHL2 A0A084VMX7 A0A182QFZ3 A0A1B6MIT4 A0A0K8U458 A0A1A9UH41 A0A1A9XT66 A0A1A9ZFA8 A0A1B0BN40 A0A0L0BMK7 A0A1A9W5K5 A0A0V0G7S9 A0A224XAM9 A0A1B6J7N9 F4WYE3 A0A1B6JM06 A0A1B6HC48 A0A195EUR8 N6T2J8 A0A195ECR6 A0A195B0A8 A0A158NXP7 A0A0A9YKT4 W8B721 A0A0A1XNN5 A0A0A1XN77 E0VII0 B4M9H8 A0A0K8UK01 A0A3B0JJ67 B4GSB9 Q29KF6 A0A0M4EBL4 B4I2S1 B4Q8M2 Q9VQI2 B4NXA0 B3NA79 B4N000 B4KH11 A0A1B6C8I6 A0A1B6GD71
PDB
2DDU
E-value=0.00267859,
Score=91
Ontologies
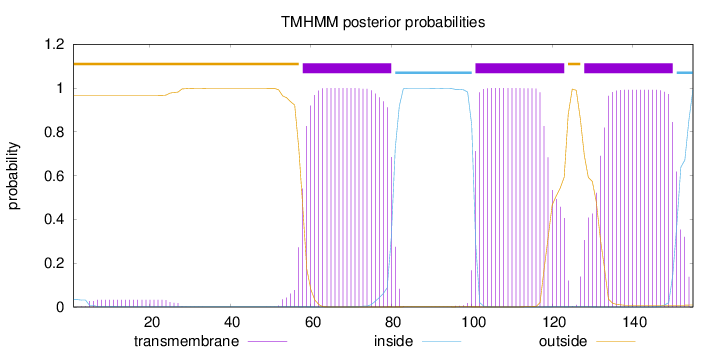
Topology
Length:
155
Number of predicted TMHs:
3
Exp number of AAs in TMHs:
65.03824
Exp number, first 60 AAs:
3.49715
Total prob of N-in:
0.03409
outside
1 - 57
TMhelix
58 - 80
inside
81 - 100
TMhelix
101 - 123
outside
124 - 127
TMhelix
128 - 150
inside
151 - 155
Population Genetic Test Statistics
Pi
211.048317
Theta
151.6103
Tajima's D
1.188836
CLR
0.124227
CSRT
0.711464426778661
Interpretation
Uncertain
