Gene
KWMTBOMO12185
Pre Gene Modal
BGIBMGA014450
Annotation
PREDICTED:_uncharacterized_protein_LOC101742687_isoform_X2_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 3.333
Sequence
CDS
ATGTCAAGCTATGCTGGTCCTCCTCCTATCGTCGTTTTCCTCATTACTGAGGGTCGTGACTCCCCCGTTGCATCAGTTTCTCCCGTACGATTTCCCTCCATCTGCTGCGATCCTTATGGGGGGTTTACACGGGTGTTTCACCTTCGCAACATCCGCTGGGAGAGTCCAAGGTTCCTGTCCAAATCGTTCCTGGCCTTCGTTACCTTCAACCTCATCAACTACTCGTTGTTGGCAGCCGAGGTCCTTACGACGAACATCGCTGATACTTCTGCCGAGAAGAGGACCTTCTACCAGCACATATTCAATGGTTGTTACGCCGTTCTACTCTTCATCGTTGTCGTGTTCTTCCTCATCTACGGTGTAGAAGTATTCTTTAAGCTCCGCGGTGAGTTCCTGAAGGAGACCAAAGTATGCACCATAATATCGCCCCGTCCCGGAACCTCAGCCGACGGAGATGAAGCCCAGGATACGCTTGTCACTAAACAGGGTGCCGGTGGTGTCCCGCAACTGGCACGACCTGTGTTCCGGCTCGCCGAGATCGGGGTCGCAGTATGGTTCCCGTGCGCGCTGTGGAACTCGATGGCGCCCGAGCGGCTCTGGCTCCTGAACCCGCGCCGGCTGCTCGCCAGGCAGCTGGACGACGCCAAGCTCGCCGACCTGCTCGCGCAGCACCACCACGATGCCAAGAACGTGGAGAGTGACTCGCTCGTGGACAGCGTCGGCTCGAGCCGGGATTGCTGGATTTGCTACGACAGTTCCCGGGCGGAGCCCCTCATCAGGCCGTGTCGCTGCACCGGCGACGTGTCCGCCGTGCATCATGACTGCCTGAGCAGGTGGCTCGTCGAGAGTGGAGTGGGAGCGCGGGTCACGTGGTCGCACTGGCGGGCACCGCGGCCGCCGTCACCGGCGCTCACCGGTGCCGCGGCCGCCCTGGGCCTCGCGCAGCTCTACCCCTCCTCCCTCGTGCGCGTGCTCGCCGCCGGCGCCGCGCTGCTCGTCTGCTACGTCGCGCTCAGATTCCTGGGCACGAACACGGTGTCGGCGTACCACCGGGCCCGCGTCTACGGCCTGAAGATCCTGACGGAGCCCTTCGCCGACGTCCAGGACTCGGAGCAGGGTCAGCTGTCGACCATCAGCAGGACGGTGACCGTCGAGATCGCGCCGAAGGACGTCCTCGACCAAGCGCTCAAGGGCGAAGCGAAATAG
Protein
MSSYAGPPPIVVFLITEGRDSPVASVSPVRFPSICCDPYGGFTRVFHLRNIRWESPRFLSKSFLAFVTFNLINYSLLAAEVLTTNIADTSAEKRTFYQHIFNGCYAVLLFIVVVFFLIYGVEVFFKLRGEFLKETKVCTIISPRPGTSADGDEAQDTLVTKQGAGGVPQLARPVFRLAEIGVAVWFPCALWNSMAPERLWLLNPRRLLARQLDDAKLADLLAQHHHDAKNVESDSLVDSVGSSRDCWICYDSSRAEPLIRPCRCTGDVSAVHHDCLSRWLVESGVGARVTWSHWRAPRPPSPALTGAAAALGLAQLYPSSLVRVLAAGAALLVCYVALRFLGTNTVSAYHRARVYGLKILTEPFADVQDSEQGQLSTISRTVTVEIAPKDVLDQALKGEAK
Summary
Uniprot
A0A3S2NLL7
A0A194QL88
A0A1E1VY05
A0A194QQX4
A0A2J7QQS1
A0A232EQV4
+ More
A0A026WJH2 A0A2J7QQT6 A0A195ECR6 F4WYE3 A0A067QZ96 A0A151INS8 E2AXV2 A0A195B0A8 A0A158NXP7 T1P9F6 T1PDL3 A0A151WPN3 B4I2S1 B4Q8M2 Q9VQI2 E1ZYP8 W8B721 A0A0Q9VYT1 A0A195EUR8 A0A0A1XN77 B4NXA0 B3MLA9 A0A3B0JJ67 B4KH11 B4N000 A0A1W4V775 B4GSB9 Q29KF6 B3NA79 B4JCE1 D6WSX5 E2BS63 B4M9H8 E0VII0 A0A1B0GGF9 U5ENJ3 A0A1A9W5K5 N6T2J8 A0A1Y1K655 U4UU81 A0A087ZQA5 A0A2A3EHR1 V9IJG0 A0A0N0BHI5 A0A0C9QZD4 A0A336LTJ9 E9FUJ6 T1HMY7 A0A0A9YKT4 A0A0V0G7S9 A0A224XAM9
A0A026WJH2 A0A2J7QQT6 A0A195ECR6 F4WYE3 A0A067QZ96 A0A151INS8 E2AXV2 A0A195B0A8 A0A158NXP7 T1P9F6 T1PDL3 A0A151WPN3 B4I2S1 B4Q8M2 Q9VQI2 E1ZYP8 W8B721 A0A0Q9VYT1 A0A195EUR8 A0A0A1XN77 B4NXA0 B3MLA9 A0A3B0JJ67 B4KH11 B4N000 A0A1W4V775 B4GSB9 Q29KF6 B3NA79 B4JCE1 D6WSX5 E2BS63 B4M9H8 E0VII0 A0A1B0GGF9 U5ENJ3 A0A1A9W5K5 N6T2J8 A0A1Y1K655 U4UU81 A0A087ZQA5 A0A2A3EHR1 V9IJG0 A0A0N0BHI5 A0A0C9QZD4 A0A336LTJ9 E9FUJ6 T1HMY7 A0A0A9YKT4 A0A0V0G7S9 A0A224XAM9
Pubmed
26354079
28648823
24508170
30249741
21719571
24845553
+ More
20798317 21347285 25315136 17994087 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24495485 25830018 17550304 18057021 15632085 23185243 18362917 19820115 20566863 23537049 28004739 21292972 25401762 26823975
20798317 21347285 25315136 17994087 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24495485 25830018 17550304 18057021 15632085 23185243 18362917 19820115 20566863 23537049 28004739 21292972 25401762 26823975
EMBL
RSAL01000042
RVE50782.1
KQ459053
KPJ04206.1
GDQN01011454
JAT79600.1
+ More
KQ461181 KPJ07749.1 NEVH01012083 PNF30922.1 NNAY01002716 OXU20712.1 KK107168 QOIP01000001 EZA56197.1 RLU26724.1 PNF30927.1 KQ979074 KYN23015.1 GL888440 EGI60795.1 KK853083 KDR11659.1 KQ976914 KYN07095.1 GL443736 EFN61716.1 KQ976692 KYM77725.1 ADTU01003275 ADTU01003276 KA644795 AFP59424.1 KA646225 AFP60854.1 KQ982851 KYQ49862.1 CH480820 EDW54066.1 CM000361 CM002910 EDX03539.1 KMY87764.1 AE014134 BT006012 AAF51189.1 AAO74695.1 GL435242 EFN73588.1 GAMC01013674 GAMC01013672 JAB92883.1 CH940654 KRF77978.1 KQ981958 KYN32005.1 GBXI01013344 GBXI01003451 GBXI01001890 JAD00948.1 JAD10841.1 JAD12402.1 CM000157 EDW87457.1 KRJ97161.1 KRJ97162.1 CH902620 EDV30698.1 KPU72993.1 KPU72994.1 OUUW01000006 SPP82397.1 CH933807 EDW12222.1 KRG03157.1 CH963920 EDW77935.1 CH479189 EDW25666.1 CH379061 EAL33219.1 KRT04816.1 KRT04817.1 CH954177 EDV57542.1 KQS70014.1 KQS70015.1 CH916368 EDW03095.1 KQ971352 EFA06336.1 GL450151 EFN81455.1 EDW57854.1 KRF77977.1 KRF77979.1 DS235200 EEB13186.1 CCAG010011257 GANO01004053 JAB55818.1 APGK01054741 APGK01054742 APGK01054743 APGK01054744 APGK01054745 APGK01054746 APGK01054747 KB741253 ENN71778.1 GEZM01097310 JAV54277.1 KB632372 ERL93751.1 KZ288254 PBC30726.1 JR050389 AEY61263.1 KQ435750 KOX76311.1 GBYB01006077 JAG75844.1 UFQT01000101 SSX19993.1 GL732525 EFX88912.1 ACPB03011978 GBHO01011363 GBRD01003907 GDHC01015330 JAG32241.1 JAG61914.1 JAQ03299.1 GECL01002072 JAP04052.1 GFTR01007031 JAW09395.1
KQ461181 KPJ07749.1 NEVH01012083 PNF30922.1 NNAY01002716 OXU20712.1 KK107168 QOIP01000001 EZA56197.1 RLU26724.1 PNF30927.1 KQ979074 KYN23015.1 GL888440 EGI60795.1 KK853083 KDR11659.1 KQ976914 KYN07095.1 GL443736 EFN61716.1 KQ976692 KYM77725.1 ADTU01003275 ADTU01003276 KA644795 AFP59424.1 KA646225 AFP60854.1 KQ982851 KYQ49862.1 CH480820 EDW54066.1 CM000361 CM002910 EDX03539.1 KMY87764.1 AE014134 BT006012 AAF51189.1 AAO74695.1 GL435242 EFN73588.1 GAMC01013674 GAMC01013672 JAB92883.1 CH940654 KRF77978.1 KQ981958 KYN32005.1 GBXI01013344 GBXI01003451 GBXI01001890 JAD00948.1 JAD10841.1 JAD12402.1 CM000157 EDW87457.1 KRJ97161.1 KRJ97162.1 CH902620 EDV30698.1 KPU72993.1 KPU72994.1 OUUW01000006 SPP82397.1 CH933807 EDW12222.1 KRG03157.1 CH963920 EDW77935.1 CH479189 EDW25666.1 CH379061 EAL33219.1 KRT04816.1 KRT04817.1 CH954177 EDV57542.1 KQS70014.1 KQS70015.1 CH916368 EDW03095.1 KQ971352 EFA06336.1 GL450151 EFN81455.1 EDW57854.1 KRF77977.1 KRF77979.1 DS235200 EEB13186.1 CCAG010011257 GANO01004053 JAB55818.1 APGK01054741 APGK01054742 APGK01054743 APGK01054744 APGK01054745 APGK01054746 APGK01054747 KB741253 ENN71778.1 GEZM01097310 JAV54277.1 KB632372 ERL93751.1 KZ288254 PBC30726.1 JR050389 AEY61263.1 KQ435750 KOX76311.1 GBYB01006077 JAG75844.1 UFQT01000101 SSX19993.1 GL732525 EFX88912.1 ACPB03011978 GBHO01011363 GBRD01003907 GDHC01015330 JAG32241.1 JAG61914.1 JAQ03299.1 GECL01002072 JAP04052.1 GFTR01007031 JAW09395.1
Proteomes
UP000283053
UP000053268
UP000053240
UP000235965
UP000215335
UP000053097
+ More
UP000279307 UP000078492 UP000007755 UP000027135 UP000078542 UP000000311 UP000078540 UP000005205 UP000095301 UP000075809 UP000001292 UP000000304 UP000000803 UP000008792 UP000078541 UP000002282 UP000007801 UP000268350 UP000009192 UP000007798 UP000192221 UP000008744 UP000001819 UP000008711 UP000001070 UP000007266 UP000008237 UP000009046 UP000092444 UP000091820 UP000019118 UP000030742 UP000005203 UP000242457 UP000053105 UP000000305 UP000015103
UP000279307 UP000078492 UP000007755 UP000027135 UP000078542 UP000000311 UP000078540 UP000005205 UP000095301 UP000075809 UP000001292 UP000000304 UP000000803 UP000008792 UP000078541 UP000002282 UP000007801 UP000268350 UP000009192 UP000007798 UP000192221 UP000008744 UP000001819 UP000008711 UP000001070 UP000007266 UP000008237 UP000009046 UP000092444 UP000091820 UP000019118 UP000030742 UP000005203 UP000242457 UP000053105 UP000000305 UP000015103
PRIDE
Interpro
Gene 3D
ProteinModelPortal
A0A3S2NLL7
A0A194QL88
A0A1E1VY05
A0A194QQX4
A0A2J7QQS1
A0A232EQV4
+ More
A0A026WJH2 A0A2J7QQT6 A0A195ECR6 F4WYE3 A0A067QZ96 A0A151INS8 E2AXV2 A0A195B0A8 A0A158NXP7 T1P9F6 T1PDL3 A0A151WPN3 B4I2S1 B4Q8M2 Q9VQI2 E1ZYP8 W8B721 A0A0Q9VYT1 A0A195EUR8 A0A0A1XN77 B4NXA0 B3MLA9 A0A3B0JJ67 B4KH11 B4N000 A0A1W4V775 B4GSB9 Q29KF6 B3NA79 B4JCE1 D6WSX5 E2BS63 B4M9H8 E0VII0 A0A1B0GGF9 U5ENJ3 A0A1A9W5K5 N6T2J8 A0A1Y1K655 U4UU81 A0A087ZQA5 A0A2A3EHR1 V9IJG0 A0A0N0BHI5 A0A0C9QZD4 A0A336LTJ9 E9FUJ6 T1HMY7 A0A0A9YKT4 A0A0V0G7S9 A0A224XAM9
A0A026WJH2 A0A2J7QQT6 A0A195ECR6 F4WYE3 A0A067QZ96 A0A151INS8 E2AXV2 A0A195B0A8 A0A158NXP7 T1P9F6 T1PDL3 A0A151WPN3 B4I2S1 B4Q8M2 Q9VQI2 E1ZYP8 W8B721 A0A0Q9VYT1 A0A195EUR8 A0A0A1XN77 B4NXA0 B3MLA9 A0A3B0JJ67 B4KH11 B4N000 A0A1W4V775 B4GSB9 Q29KF6 B3NA79 B4JCE1 D6WSX5 E2BS63 B4M9H8 E0VII0 A0A1B0GGF9 U5ENJ3 A0A1A9W5K5 N6T2J8 A0A1Y1K655 U4UU81 A0A087ZQA5 A0A2A3EHR1 V9IJG0 A0A0N0BHI5 A0A0C9QZD4 A0A336LTJ9 E9FUJ6 T1HMY7 A0A0A9YKT4 A0A0V0G7S9 A0A224XAM9
PDB
2D8S
E-value=0.00287592,
Score=96
Ontologies
GO
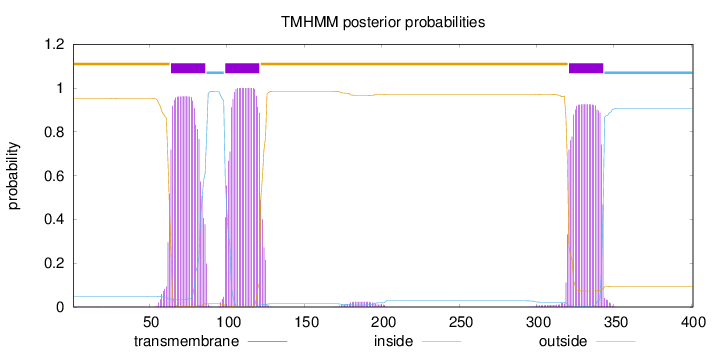
Topology
Length:
401
Number of predicted TMHs:
3
Exp number of AAs in TMHs:
64.3749200000001
Exp number, first 60 AAs:
0.29568
Total prob of N-in:
0.04800
outside
1 - 63
TMhelix
64 - 86
inside
87 - 98
TMhelix
99 - 121
outside
122 - 320
TMhelix
321 - 343
inside
344 - 401
Population Genetic Test Statistics
Pi
388.99186
Theta
139.294025
Tajima's D
-0.931011
CLR
1.62608
CSRT
0.147142642867857
Interpretation
Uncertain
