Gene
KWMTBOMO12184
Pre Gene Modal
BGIBMGA014450
Annotation
PREDICTED:_uncharacterized_protein_LOC101742687_isoform_X2_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 3.671
Sequence
CDS
ATGCTTATTGGCGATAGAAATACGCCGGTATCATACTTAGAGGCTTGCCCTGAGGAGTTGAAGAAAATATGTCAAGTTATGCTGGTCCTCCTCCTCGCGTCGTTTTCCTCATTACTGAGGGTCGTGACTCCCCCGCTGCATCAGTTTCTCCCGTACGACTTCCCTCCATCTGCTGCGATCCTTAGCTTCATGAAGGGCATTGTGGATTTGATGTTTAGAGAAAATCGTATTTGGTCCGACCATCTCGTGTTTCACCTTCGCAACATCCGCTGGGAGAGTCCAAGGTTCCTGTCCAAATCGTTCCTGGCCTTCGTTACATTCAACCTCATCAACTACTCGTTGTTGGCAGCCGAGGTCCTTACAACGAACATCGCTGATACTTCTGCCGAGAAGAGGACCTTCTACCAGCACATATTCAATGGTTGTTACGCCGTTCTACTCTTCATCGTTGTCGTGTTCTTCCTCATCTACGGAGTTGAAGTATTCTTTAAGCTCCGCGGTGAGTTCCTGAAGGAGACCAAAGTATGCACCATAATATCGCCCCGTCCCGGAACCTCAACCGACGGAGATGAAGCCCAGGATACGCTTGTCACTAAACAGGGAATTCAAACGGATTTGCTGGACGGAGACGGAACGGTGGACCCTCGGTCCGTGGACCTGTCGCAGCTGCACCAGTCGCGCGTGGGCCTGATCAGCCTCGCGCTGATGCTCATACTGATCGTCTGCTTCCTGGCTTCTGAAACCCTAAGCGAATTCTGGAAGACTAAGGTGCCGGTGGTGTCCCGCAACTGGCACGACCTGGTGTTCCGGCTCGCCGAGATCGGGGTCGCAGTATGGTTCCCGTGCGCGCTGTGGAACTCGATGGCGCCCGAGCGGCTCTGGCTCCTGAACCCGCGCCGGCTGCTCGCCAGGCAACTGGACGACGCCAAGCTCGCCGACCTGCTCGCGCAGCACCACCACGATGCCAAGACGTTGGCCGACCTCAGCCAACTGGGCTGGCGCCAGAAAGCATGGTACTTTCTCGAGTTGGCCAAATATTGGATCGTTTGGAGGTTCGCTCTTACTGTCTTTTTCAAACCTCTTCCTTTGCCTGTAAACGGGAAAGAGACACAAAACTCGGACGCTGCTAATGCGGTTGGCTCGAACACAAATTCGAACGTGGAGAGAGACTCGCTCGTGGACAGCGTCGGGTCGAGCCGCGACTGCTGGATTTGCTACGACAGCTCCCGGGCGGAGCCCCTCATCAGGCCGTGTCGCTGCACAGGCGACGTGTCCGCCGTGCATCATGACTGCCTGAGCAGGTGGCTCGTTGAGAGTGCCGCCTCGCCTGACGGCTTGAAGTGCAAAGTCTGCAACACTCCCTACATCGTCCAGGAGACCAATCGAGTGGAGTGGGAGCGCGGGTTCACGTGGTCGCACTGGGCGGGCACCGCGGCCGCCGTCACCGCGCTCACCGGCGCCGCGGCGGGCGCCTGGGCCCTCGCGCAGCTCTACCCCTCCTCCCTCGTACGCGTGCTCGCCGCCGGCGCCGCGCTGCTCGTCTGCTACGTCGCGCTCAGATTCCTGGGCACGAACACGGTGTCGGCGTACCACCGGGCCCGCGTCTACGGCCTGAAGATCCTGACGGAGCCCTTCGCCGACGTCCAGGACTCGGAGCAGGGTCAGCTGTCGACCATCAGCAGGACGGTGACCGTCGAGATCGCACCGAAGGACGTCCTCGACCAAGCGCTCAAGGGCGAAGCGAAATAG
Protein
MLIGDRNTPVSYLEACPEELKKICQVMLVLLLASFSSLLRVVTPPLHQFLPYDFPPSAAILSFMKGIVDLMFRENRIWSDHLVFHLRNIRWESPRFLSKSFLAFVTFNLINYSLLAAEVLTTNIADTSAEKRTFYQHIFNGCYAVLLFIVVVFFLIYGVEVFFKLRGEFLKETKVCTIISPRPGTSTDGDEAQDTLVTKQGIQTDLLDGDGTVDPRSVDLSQLHQSRVGLISLALMLILIVCFLASETLSEFWKTKVPVVSRNWHDLVFRLAEIGVAVWFPCALWNSMAPERLWLLNPRRLLARQLDDAKLADLLAQHHHDAKTLADLSQLGWRQKAWYFLELAKYWIVWRFALTVFFKPLPLPVNGKETQNSDAANAVGSNTNSNVERDSLVDSVGSSRDCWICYDSSRAEPLIRPCRCTGDVSAVHHDCLSRWLVESAASPDGLKCKVCNTPYIVQETNRVEWERGFTWSHWAGTAAAVTALTGAAAGAWALAQLYPSSLVRVLAAGAALLVCYVALRFLGTNTVSAYHRARVYGLKILTEPFADVQDSEQGQLSTISRTVTVEIAPKDVLDQALKGEAK
Summary
Uniprot
Pubmed
EMBL
NWSH01002568
PCG67959.1
GDIQ01245850
GDIQ01209105
GDIQ01209104
GDIQ01191544
+ More
GDIQ01187615 GDIQ01166811 GDIQ01166810 GDIQ01149577 GDIQ01085806 JAN08931.1 GDIQ01014549 JAN80188.1 LRGB01000868 KZS15587.1 GDIQ01227899 JAK23826.1 GDIP01144877 JAJ78525.1 GDIQ01227898 JAK23827.1 GEFM01006437 JAP69359.1 GFAC01004575 JAT94613.1
GDIQ01187615 GDIQ01166811 GDIQ01166810 GDIQ01149577 GDIQ01085806 JAN08931.1 GDIQ01014549 JAN80188.1 LRGB01000868 KZS15587.1 GDIQ01227899 JAK23826.1 GDIP01144877 JAJ78525.1 GDIQ01227898 JAK23827.1 GEFM01006437 JAP69359.1 GFAC01004575 JAT94613.1
Proteomes
Pfam
PF12906 RINGv
Gene 3D
ProteinModelPortal
PDB
2D8S
E-value=0.000104324,
Score=110
Ontologies
GO
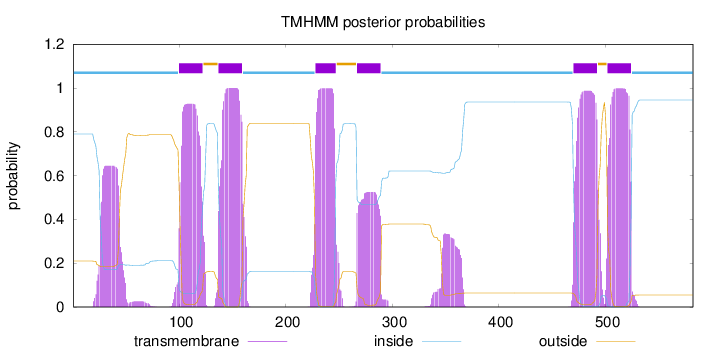
Topology
Length:
582
Number of predicted TMHs:
6
Exp number of AAs in TMHs:
139.4621
Exp number, first 60 AAs:
13.39902
Total prob of N-in:
0.79043
POSSIBLE N-term signal
sequence
inside
1 - 99
TMhelix
100 - 122
outside
123 - 136
TMhelix
137 - 159
inside
160 - 227
TMhelix
228 - 247
outside
248 - 266
TMhelix
267 - 289
inside
290 - 469
TMhelix
470 - 492
outside
493 - 501
TMhelix
502 - 524
inside
525 - 582
Population Genetic Test Statistics
Pi
328.071808
Theta
17.662597
Tajima's D
2.895164
CLR
0.247022
CSRT
0.974651267436628
Interpretation
Uncertain
