Gene
KWMTBOMO12181
Pre Gene Modal
BGIBMGA004228
Annotation
PREDICTED:_NF-kappa-B_inhibitor-interacting_Ras-like_protein_isoform_X1_[Papilio_polytes]
Full name
NF-kappa-B inhibitor-interacting Ras-like protein
Location in the cell
Cytoplasmic Reliability : 2.024 Nuclear Reliability : 1.491
Sequence
CDS
ATGGGTAAAACAAGCAAAGTTGTTGTATGTGGCATGAAGGGAGTCGGTAAAACAGCTATCCTAGAGCAGTTAATTTACGGCAATGTAAATTTGAAATCATGTTTTTATCCCACGATCGAAGACATTTATGTGGCGAACATCGAAACAGATCGCGGCCCGAAAGAACGCGTCTGTTTCTACGACACAGCGGGCTTGGAGCCTCCGCTTACAGGCGAGTTTAAAGGTCCTCCGCAATCTCCTACAATGCAGCTAACGGCCAACCAAGTGCTCCAACGACACTATCTCGGTTTCTCAGAAGGTTTTGTCATGGTGTATGATACAGCGCGACCAGAATCGCTGGATGTACTCATGTATTTAAAGAAGGACATAGACAGGAACAAAGATAAGAAAGAGGTAGTAATACTAGTGATCGGTAATAGAACCGGACCAGATGATCCGAACAGCCTCGAGAACACATGCTCCAAAGCCTCGAATTGGTGTGCACGCGAGAAGGTCAGACATTTTGAGGTGAACGCAATGGATCGTCCGTCCCTATATGAACCATTCATCTTCATGACCTCGAAATTGAACCCGGTACAAAATAAGACGACCTTCCCGCAACTAAGCACGTTGAGTAAGCTCACCCAGAAGAACAATAAAAGTGAAGCCATGTAA
Protein
MGKTSKVVVCGMKGVGKTAILEQLIYGNVNLKSCFYPTIEDIYVANIETDRGPKERVCFYDTAGLEPPLTGEFKGPPQSPTMQLTANQVLQRHYLGFSEGFVMVYDTARPESLDVLMYLKKDIDRNKDKKEVVILVIGNRTGPDDPNSLENTCSKASNWCAREKVRHFEVNAMDRPSLYEPFIFMTSKLNPVQNKTTFPQLSTLSKLTQKNNKSEAM
Summary
Description
Atypical Ras-like protein that may act as a regulator of NF-kappa-B activity, possibly by preventing the degradation of NF-kappa-B inhibitor cactus.
Subunit
Interacts with NF-kappa-B inhibitor cactus.
Similarity
Belongs to the small GTPase superfamily. Ras family. KappaB-Ras subfamily.
Keywords
Alternative splicing
Complete proteome
GTP-binding
Nucleotide-binding
Reference proteome
Feature
chain NF-kappa-B inhibitor-interacting Ras-like protein
splice variant In isoform B.
splice variant In isoform B.
Uniprot
H9J3Y9
A0A2W1BG52
A0A2H1V4V3
A0A2A4JA88
A0A194QQH1
A0A212EXM3
+ More
A0A2A4J978 S4PCW5 A0A0L7L7F0 A0A232EKH7 K7JAA5 A0A2A3EFP6 V9IMD4 A0A087ZYU6 A0A0L7RH93 A0A0M8ZZ00 A0A154PQE3 A0A310SRP6 A0A067RL41 E2C0E3 A0A026WAP8 A0A2J7PSG3 E2AVR9 A0A0J7K9R9 A0A158NYK3 A0A151JWH2 F4X316 A0A151WQB7 A0A195AZ93 A0A151JND8 A0A195D5L3 A0A2P8XFK6 E9JDE3 A0A0T6AW56 A0A139WE77 A0A1B6MFH4 J3JX19 A0A2H8TCS4 A0A0L0BVX5 A0A1B6DQR9 A0A1I8NIN3 B4HQU6 B3N9Z4 A0A1A9X4A7 B4QEA1 B7YZS7 Q9V4L4 A0A1A9UER9 B4P1P3 B4LMP6 A0A1W4V0X9 Q1HQI5 D3TQF0 A0A1A9ZQT7 A0A0M4EBF5 A0A1A9X9E6 J9JUZ2 B3MDQ3 A0A1B0G2V2 B4KND2 W8C2Z0 A0A182HBU6 Q9V4L4-2 A0A0K8VAZ6 A0A182TXC3 A0A1Y1LBT5 A0A0K8TLV8 A0A3B0J2K3 B0W2P0 B4J6P3 A0A182HYA6 A0A0A1XGF2 A0A2S2NNH5 A0A182LHF2 A0A1Q3EXF0 A0A034WPD0 B5DUR5 B4GIM6 A0A1S3D464 A0A1Q3EXE6 T1H660 Q7QHP2 A0A1Q3EXW1 A0A182UTH3 A0A084VD30 A0A1I8NXT3 A0A182P0R0 A0A182JB22 A0A182WYS8 A0A336MLW8 B4MR67 A0A182KEA4 B4NUZ0 A0A1B0DCY3 A0A1L8DLZ1 A0A1B0GH24 C3ZPP9
A0A2A4J978 S4PCW5 A0A0L7L7F0 A0A232EKH7 K7JAA5 A0A2A3EFP6 V9IMD4 A0A087ZYU6 A0A0L7RH93 A0A0M8ZZ00 A0A154PQE3 A0A310SRP6 A0A067RL41 E2C0E3 A0A026WAP8 A0A2J7PSG3 E2AVR9 A0A0J7K9R9 A0A158NYK3 A0A151JWH2 F4X316 A0A151WQB7 A0A195AZ93 A0A151JND8 A0A195D5L3 A0A2P8XFK6 E9JDE3 A0A0T6AW56 A0A139WE77 A0A1B6MFH4 J3JX19 A0A2H8TCS4 A0A0L0BVX5 A0A1B6DQR9 A0A1I8NIN3 B4HQU6 B3N9Z4 A0A1A9X4A7 B4QEA1 B7YZS7 Q9V4L4 A0A1A9UER9 B4P1P3 B4LMP6 A0A1W4V0X9 Q1HQI5 D3TQF0 A0A1A9ZQT7 A0A0M4EBF5 A0A1A9X9E6 J9JUZ2 B3MDQ3 A0A1B0G2V2 B4KND2 W8C2Z0 A0A182HBU6 Q9V4L4-2 A0A0K8VAZ6 A0A182TXC3 A0A1Y1LBT5 A0A0K8TLV8 A0A3B0J2K3 B0W2P0 B4J6P3 A0A182HYA6 A0A0A1XGF2 A0A2S2NNH5 A0A182LHF2 A0A1Q3EXF0 A0A034WPD0 B5DUR5 B4GIM6 A0A1S3D464 A0A1Q3EXE6 T1H660 Q7QHP2 A0A1Q3EXW1 A0A182UTH3 A0A084VD30 A0A1I8NXT3 A0A182P0R0 A0A182JB22 A0A182WYS8 A0A336MLW8 B4MR67 A0A182KEA4 B4NUZ0 A0A1B0DCY3 A0A1L8DLZ1 A0A1B0GH24 C3ZPP9
Pubmed
19121390
28756777
26354079
22118469
23622113
26227816
+ More
28648823 20075255 24845553 20798317 24508170 30249741 21347285 21719571 29403074 21282665 18362917 19820115 22516182 23537049 26108605 25315136 17994087 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 10657303 12537569 17550304 17204158 17510324 20353571 24495485 26483478 28004739 26369729 25830018 20966253 25348373 15632085 12364791 14747013 17210077 24438588 18563158
28648823 20075255 24845553 20798317 24508170 30249741 21347285 21719571 29403074 21282665 18362917 19820115 22516182 23537049 26108605 25315136 17994087 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 10657303 12537569 17550304 17204158 17510324 20353571 24495485 26483478 28004739 26369729 25830018 20966253 25348373 15632085 12364791 14747013 17210077 24438588 18563158
EMBL
BABH01040242
BABH01040243
KZ150117
PZC73261.1
ODYU01000663
SOQ35829.1
+ More
NWSH01002285 PCG68676.1 KQ461181 KPJ07747.1 AGBW02011714 OWR46240.1 PCG68677.1 GAIX01003926 JAA88634.1 JTDY01002495 KOB71295.1 NNAY01003790 OXU18866.1 AAZX01002420 KZ288269 PBC29976.1 JR053624 AEY61977.1 KQ414596 KOC70086.1 KQ435809 KOX72966.1 KQ435037 KZC14102.1 KQ759870 OAD62447.1 KK852611 KDR20254.1 GL451770 EFN78602.1 KK107295 QOIP01000003 EZA53058.1 RLU24417.1 NEVH01021928 PNF19271.1 GL443183 EFN62478.1 LBMM01010920 KMQ87153.1 ADTU01003975 KQ981647 KYN38655.1 GL888608 EGI59116.1 KQ982843 KYQ49998.1 KQ976701 KYM77355.1 KQ978848 KYN27969.1 KQ976818 KYN08141.1 PYGN01002316 PSN30779.1 GL771866 EFZ09170.1 LJIG01022664 KRT79370.1 KQ971354 KYB26253.1 GEBQ01005270 GEBQ01002419 JAT34707.1 JAT37558.1 APGK01024144 BT127787 KB740539 KB632197 AEE62749.1 ENN80443.1 ERL90028.1 GFXV01000106 MBW11911.1 JRES01001254 KNC24192.1 GEDC01009264 JAS28034.1 CH480816 EDW46756.1 CH954177 EDV59690.1 CM000362 CM002911 EDX06010.1 KMY91974.1 AE013599 ACL83067.1 AGB93303.1 AF229841 AY061054 BT003452 CM000157 EDW89179.1 CH940648 EDW60987.1 DQ440459 CH477283 ABF18492.1 EAT44856.1 EZ423652 ADD19928.1 CP012524 ALC40852.1 ABLF02039936 CH902619 EDV36438.1 CCAG010021272 CH933808 EDW09985.1 GAMC01000248 JAC06308.1 JXUM01032435 KQ560955 KXJ80224.1 GDHF01016293 JAI36021.1 GEZM01061575 GEZM01061574 GEZM01061573 JAV70358.1 GDAI01002683 JAI14920.1 OUUW01000001 SPP75674.1 DS231827 EDS29348.1 CH916367 EDW00946.1 APCN01004511 GBXI01004297 JAD09995.1 GGMR01006131 MBY18750.1 GFDL01015060 JAV19985.1 GAKP01001511 JAC57441.1 CH674335 EDY71634.1 CH479183 EDW36346.1 GFDL01015061 JAV19984.1 AAAB01008816 EAA05174.3 GFDL01014902 JAV20143.1 ATLV01011135 KE524643 KFB35874.1 UFQS01001683 UFQT01000834 UFQT01001683 SSX11848.1 SSX27492.1 SSX31412.1 CH963849 EDW74606.1 CH984740 EDX16085.1 AJVK01014283 GFDF01006689 JAV07395.1 AJWK01003897 GG666659 EEN45470.1
NWSH01002285 PCG68676.1 KQ461181 KPJ07747.1 AGBW02011714 OWR46240.1 PCG68677.1 GAIX01003926 JAA88634.1 JTDY01002495 KOB71295.1 NNAY01003790 OXU18866.1 AAZX01002420 KZ288269 PBC29976.1 JR053624 AEY61977.1 KQ414596 KOC70086.1 KQ435809 KOX72966.1 KQ435037 KZC14102.1 KQ759870 OAD62447.1 KK852611 KDR20254.1 GL451770 EFN78602.1 KK107295 QOIP01000003 EZA53058.1 RLU24417.1 NEVH01021928 PNF19271.1 GL443183 EFN62478.1 LBMM01010920 KMQ87153.1 ADTU01003975 KQ981647 KYN38655.1 GL888608 EGI59116.1 KQ982843 KYQ49998.1 KQ976701 KYM77355.1 KQ978848 KYN27969.1 KQ976818 KYN08141.1 PYGN01002316 PSN30779.1 GL771866 EFZ09170.1 LJIG01022664 KRT79370.1 KQ971354 KYB26253.1 GEBQ01005270 GEBQ01002419 JAT34707.1 JAT37558.1 APGK01024144 BT127787 KB740539 KB632197 AEE62749.1 ENN80443.1 ERL90028.1 GFXV01000106 MBW11911.1 JRES01001254 KNC24192.1 GEDC01009264 JAS28034.1 CH480816 EDW46756.1 CH954177 EDV59690.1 CM000362 CM002911 EDX06010.1 KMY91974.1 AE013599 ACL83067.1 AGB93303.1 AF229841 AY061054 BT003452 CM000157 EDW89179.1 CH940648 EDW60987.1 DQ440459 CH477283 ABF18492.1 EAT44856.1 EZ423652 ADD19928.1 CP012524 ALC40852.1 ABLF02039936 CH902619 EDV36438.1 CCAG010021272 CH933808 EDW09985.1 GAMC01000248 JAC06308.1 JXUM01032435 KQ560955 KXJ80224.1 GDHF01016293 JAI36021.1 GEZM01061575 GEZM01061574 GEZM01061573 JAV70358.1 GDAI01002683 JAI14920.1 OUUW01000001 SPP75674.1 DS231827 EDS29348.1 CH916367 EDW00946.1 APCN01004511 GBXI01004297 JAD09995.1 GGMR01006131 MBY18750.1 GFDL01015060 JAV19985.1 GAKP01001511 JAC57441.1 CH674335 EDY71634.1 CH479183 EDW36346.1 GFDL01015061 JAV19984.1 AAAB01008816 EAA05174.3 GFDL01014902 JAV20143.1 ATLV01011135 KE524643 KFB35874.1 UFQS01001683 UFQT01000834 UFQT01001683 SSX11848.1 SSX27492.1 SSX31412.1 CH963849 EDW74606.1 CH984740 EDX16085.1 AJVK01014283 GFDF01006689 JAV07395.1 AJWK01003897 GG666659 EEN45470.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000007151
UP000037510
UP000215335
+ More
UP000002358 UP000242457 UP000005203 UP000053825 UP000053105 UP000076502 UP000027135 UP000008237 UP000053097 UP000279307 UP000235965 UP000000311 UP000036403 UP000005205 UP000078541 UP000007755 UP000075809 UP000078540 UP000078492 UP000078542 UP000245037 UP000007266 UP000019118 UP000030742 UP000037069 UP000095301 UP000001292 UP000008711 UP000091820 UP000000304 UP000000803 UP000078200 UP000002282 UP000008792 UP000192221 UP000008820 UP000092445 UP000092553 UP000092443 UP000007819 UP000007801 UP000092444 UP000009192 UP000069940 UP000249989 UP000075902 UP000268350 UP000002320 UP000001070 UP000075840 UP000075882 UP000001819 UP000008744 UP000079169 UP000015102 UP000007062 UP000075903 UP000030765 UP000095300 UP000075885 UP000075880 UP000076407 UP000007798 UP000075881 UP000092462 UP000092461 UP000001554
UP000002358 UP000242457 UP000005203 UP000053825 UP000053105 UP000076502 UP000027135 UP000008237 UP000053097 UP000279307 UP000235965 UP000000311 UP000036403 UP000005205 UP000078541 UP000007755 UP000075809 UP000078540 UP000078492 UP000078542 UP000245037 UP000007266 UP000019118 UP000030742 UP000037069 UP000095301 UP000001292 UP000008711 UP000091820 UP000000304 UP000000803 UP000078200 UP000002282 UP000008792 UP000192221 UP000008820 UP000092445 UP000092553 UP000092443 UP000007819 UP000007801 UP000092444 UP000009192 UP000069940 UP000249989 UP000075902 UP000268350 UP000002320 UP000001070 UP000075840 UP000075882 UP000001819 UP000008744 UP000079169 UP000015102 UP000007062 UP000075903 UP000030765 UP000095300 UP000075885 UP000075880 UP000076407 UP000007798 UP000075881 UP000092462 UP000092461 UP000001554
Pfam
PF00071 Ras
Interpro
SUPFAM
SSF52540
SSF52540
ProteinModelPortal
H9J3Y9
A0A2W1BG52
A0A2H1V4V3
A0A2A4JA88
A0A194QQH1
A0A212EXM3
+ More
A0A2A4J978 S4PCW5 A0A0L7L7F0 A0A232EKH7 K7JAA5 A0A2A3EFP6 V9IMD4 A0A087ZYU6 A0A0L7RH93 A0A0M8ZZ00 A0A154PQE3 A0A310SRP6 A0A067RL41 E2C0E3 A0A026WAP8 A0A2J7PSG3 E2AVR9 A0A0J7K9R9 A0A158NYK3 A0A151JWH2 F4X316 A0A151WQB7 A0A195AZ93 A0A151JND8 A0A195D5L3 A0A2P8XFK6 E9JDE3 A0A0T6AW56 A0A139WE77 A0A1B6MFH4 J3JX19 A0A2H8TCS4 A0A0L0BVX5 A0A1B6DQR9 A0A1I8NIN3 B4HQU6 B3N9Z4 A0A1A9X4A7 B4QEA1 B7YZS7 Q9V4L4 A0A1A9UER9 B4P1P3 B4LMP6 A0A1W4V0X9 Q1HQI5 D3TQF0 A0A1A9ZQT7 A0A0M4EBF5 A0A1A9X9E6 J9JUZ2 B3MDQ3 A0A1B0G2V2 B4KND2 W8C2Z0 A0A182HBU6 Q9V4L4-2 A0A0K8VAZ6 A0A182TXC3 A0A1Y1LBT5 A0A0K8TLV8 A0A3B0J2K3 B0W2P0 B4J6P3 A0A182HYA6 A0A0A1XGF2 A0A2S2NNH5 A0A182LHF2 A0A1Q3EXF0 A0A034WPD0 B5DUR5 B4GIM6 A0A1S3D464 A0A1Q3EXE6 T1H660 Q7QHP2 A0A1Q3EXW1 A0A182UTH3 A0A084VD30 A0A1I8NXT3 A0A182P0R0 A0A182JB22 A0A182WYS8 A0A336MLW8 B4MR67 A0A182KEA4 B4NUZ0 A0A1B0DCY3 A0A1L8DLZ1 A0A1B0GH24 C3ZPP9
A0A2A4J978 S4PCW5 A0A0L7L7F0 A0A232EKH7 K7JAA5 A0A2A3EFP6 V9IMD4 A0A087ZYU6 A0A0L7RH93 A0A0M8ZZ00 A0A154PQE3 A0A310SRP6 A0A067RL41 E2C0E3 A0A026WAP8 A0A2J7PSG3 E2AVR9 A0A0J7K9R9 A0A158NYK3 A0A151JWH2 F4X316 A0A151WQB7 A0A195AZ93 A0A151JND8 A0A195D5L3 A0A2P8XFK6 E9JDE3 A0A0T6AW56 A0A139WE77 A0A1B6MFH4 J3JX19 A0A2H8TCS4 A0A0L0BVX5 A0A1B6DQR9 A0A1I8NIN3 B4HQU6 B3N9Z4 A0A1A9X4A7 B4QEA1 B7YZS7 Q9V4L4 A0A1A9UER9 B4P1P3 B4LMP6 A0A1W4V0X9 Q1HQI5 D3TQF0 A0A1A9ZQT7 A0A0M4EBF5 A0A1A9X9E6 J9JUZ2 B3MDQ3 A0A1B0G2V2 B4KND2 W8C2Z0 A0A182HBU6 Q9V4L4-2 A0A0K8VAZ6 A0A182TXC3 A0A1Y1LBT5 A0A0K8TLV8 A0A3B0J2K3 B0W2P0 B4J6P3 A0A182HYA6 A0A0A1XGF2 A0A2S2NNH5 A0A182LHF2 A0A1Q3EXF0 A0A034WPD0 B5DUR5 B4GIM6 A0A1S3D464 A0A1Q3EXE6 T1H660 Q7QHP2 A0A1Q3EXW1 A0A182UTH3 A0A084VD30 A0A1I8NXT3 A0A182P0R0 A0A182JB22 A0A182WYS8 A0A336MLW8 B4MR67 A0A182KEA4 B4NUZ0 A0A1B0DCY3 A0A1L8DLZ1 A0A1B0GH24 C3ZPP9
PDB
3KUC
E-value=0.000257131,
Score=102
Ontologies
GO
PANTHER
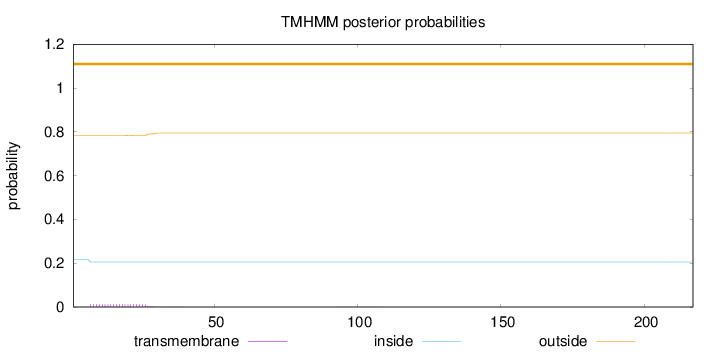
Topology
Length:
217
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.2561
Exp number, first 60 AAs:
0.25261
Total prob of N-in:
0.21723
outside
1 - 217
Population Genetic Test Statistics
Pi
220.984687
Theta
138.569983
Tajima's D
1.407084
CLR
0.392255
CSRT
0.762261886905655
Interpretation
Uncertain
