Pre Gene Modal
BGIBMGA004218
Annotation
PREDICTED:_dolichyl-phosphate_mannosyltransferase_isoform_X1_[Bombyx_mori]
Full name
Dolichol-phosphate mannosyltransferase subunit 1
Alternative Name
Dolichol-phosphate mannose synthase subunit 1
Dolichyl-phosphate beta-D-mannosyltransferase subunit 1
Mannose-P-dolichol synthase subunit 1
Dolichyl-phosphate beta-D-mannosyltransferase subunit 1
Mannose-P-dolichol synthase subunit 1
Location in the cell
Cytoplasmic Reliability : 1.018 PlasmaMembrane Reliability : 1.525
Sequence
CDS
ATGGAGGTGGATTCGGGATTGATAAAAAGAGATAAATACTCAATTTTACTACCAACTTACAATGAACGAGAAAACCTCCCTATAATAATATGGCTAATAATTAAATATTTAGACGAAAGCGGTGTCGATTACGAAGTGATCATAATAGATGATGGAAGTCCTGATGGGACTTCAGAAGTGGCGCGTCAGCTGCAGAAGCTCTATGGCTCCTCTAAAATAGTGCTTCGACCTCGCGAGATGAAGCTTGGTCTCGGAACTGCCTACATTCATGGGATACAGCAAGCTTCTGGAAATTTTATTATTATAATGGATGCGGATTTGAGCCATCATCCAAAGTTCATTCCAGAGTTCATCAAACTCCAATTAAAATATGATTACGATATTGTCTCTGGTACTCGTTACAAGGGGAGTGGTGGAGTCTACGGCTGGGACTTCAAGCGTAAGCTCATATCTAGGGGAGCTAACTTTTTGACTCAGTTAATGCTGAGACCTGGAGTTTCAGATCTGACTGGCTCATTTAGATTGTACAAAAAGGAAGTTCTCGAGAAGCTTATTCTAAGCTGCGTTTCTAAAGGATATGTCTTCCAAATGGAAATGATTATCAGAGCGAGGCAATTTGATTATTCAATTGGAGAGGTGCCTATCTCATTTGTGGATCGTGTCTACGGAGAGTCTAAACTCGGTGGTTCGGAAATAGTACAATTCGCAAAAGCCCTACTTTATCTATTTGCCACAACATAA
Protein
MEVDSGLIKRDKYSILLPTYNERENLPIIIWLIIKYLDESGVDYEVIIIDDGSPDGTSEVARQLQKLYGSSKIVLRPREMKLGLGTAYIHGIQQASGNFIIIMDADLSHHPKFIPEFIKLQLKYDYDIVSGTRYKGSGGVYGWDFKRKLISRGANFLTQLMLRPGVSDLTGSFRLYKKEVLEKLILSCVSKGYVFQMEMIIRARQFDYSIGEVPISFVDRVYGESKLGGSEIVQFAKALLYLFATT
Summary
Description
Transfers mannose from GDP-mannose to dolichol monophosphate to form dolichol phosphate mannose (Dol-P-Man) which is the mannosyl donor in pathways leading to N-glycosylation, glycosyl phosphatidylinositol membrane anchoring, and O-mannosylation of proteins.
Catalytic Activity
dolichyl phosphate + GDP-alpha-D-mannose = dolichyl beta-D-mannosyl phosphate + GDP
Subunit
Component of the dolichol-phosphate mannose (DPM) synthase complex.
Component of the dolichol-phosphate mannose (DPM) synthase complex composed of DPM1, DPM2 and DPM3; in the complex interacts directly with DPM3.
Component of the dolichol-phosphate mannose (DPM) synthase complex composed of DPM1, DPM2 and DPM3; in the complex interacts directly with DPM3.
Similarity
Belongs to the glycosyltransferase 2 family.
Keywords
Acetylation
Complete proteome
Congenital disorder of glycosylation
Congenital muscular dystrophy
Disease mutation
Dystroglycanopathy
Endoplasmic reticulum
Glycosyltransferase
Phosphoprotein
Reference proteome
Transferase
Feature
chain Dolichol-phosphate mannosyltransferase subunit 1
sequence variant In CDG1E; dbSNP:rs121908583.
sequence variant In CDG1E; dbSNP:rs121908583.
Uniprot
Q1HPN8
H9J3X9
A0A194QF83
A0A194QW04
A0A212EXP8
A0A2A4JAG0
+ More
A0A2W1BDH5 A0A2H1V4Q7 D6WPH7 A0A1B6DY92 K7JAA4 A0A182Q9N6 A0A182PFI6 A0A182RHF6 A0A087ZYS0 A0A182M2V4 A0A182YDM0 Q1HR12 A0A1B6FA00 A0A154PQH2 U5EZ22 A0A182NUK2 A0A084W219 A0A182HRL4 A0A067RTV5 A0A1W4X9X5 A0A1W4XKC6 A0A182H4F8 A0A3S2MZF2 A0A182KTP6 A0A2M3Z2T1 A0A182K721 Q7Q1B7 A0A3B3DF72 A0A023EKR8 A0A1A7XDL1 W5J999 A0A2K5IC50 A0A315WAM6 A0A226F3Z6 A0A182UQE5 A0A182U246 A0A182F6A1 A0A1A8EU67 A0A286X9A8 A0A310SWQ5 A0A2K5YCV2 H9F662 A0A2K6CEJ3 A0A2K5W3B5 I0FW34 A0A2K6K3X4 A0A2K6Q8Y9 A0A2K5NX69 A0A2M4AFC4 A0A1Y1KJJ3 J9NZX5 F6RQC7 A0A3Q1H120 A0A0L7RH18 A0A1Q3FBS5 A0A3Q7TYS9 A0A2K5DXH7 A0A3P9LHR4 H2L910 A0A182JKR3 A0A3P9I4S5 M3WC50 A0A0K8TP84 A0A3P4NM98 A0A2M4BYD1 A0A1S3DTB4 I3MNC2 A0A2M4BYD0 W8BXV1 A0A1U7TG39 A0A1S3JV08 A0A3Q7MPY8 A0A1A8J8X9 A0A1A8D3D3 A0A2U3WT56 H2P2A8 A0A0S2Z4Y5 O60762 A0A2K6V131 A0A384CN11 A0A3Q7WD07 F4WW36 V9L673 A0A3Q2QPG4 A0A2K6EVD4 H3AP13 A0A1A8UCE9 A0A1U7QWB3 A0A3B4B8Y2 I3IZJ1 H0WRY7 M3ZW98 A0A2R9CPW5 H2QKK7
A0A2W1BDH5 A0A2H1V4Q7 D6WPH7 A0A1B6DY92 K7JAA4 A0A182Q9N6 A0A182PFI6 A0A182RHF6 A0A087ZYS0 A0A182M2V4 A0A182YDM0 Q1HR12 A0A1B6FA00 A0A154PQH2 U5EZ22 A0A182NUK2 A0A084W219 A0A182HRL4 A0A067RTV5 A0A1W4X9X5 A0A1W4XKC6 A0A182H4F8 A0A3S2MZF2 A0A182KTP6 A0A2M3Z2T1 A0A182K721 Q7Q1B7 A0A3B3DF72 A0A023EKR8 A0A1A7XDL1 W5J999 A0A2K5IC50 A0A315WAM6 A0A226F3Z6 A0A182UQE5 A0A182U246 A0A182F6A1 A0A1A8EU67 A0A286X9A8 A0A310SWQ5 A0A2K5YCV2 H9F662 A0A2K6CEJ3 A0A2K5W3B5 I0FW34 A0A2K6K3X4 A0A2K6Q8Y9 A0A2K5NX69 A0A2M4AFC4 A0A1Y1KJJ3 J9NZX5 F6RQC7 A0A3Q1H120 A0A0L7RH18 A0A1Q3FBS5 A0A3Q7TYS9 A0A2K5DXH7 A0A3P9LHR4 H2L910 A0A182JKR3 A0A3P9I4S5 M3WC50 A0A0K8TP84 A0A3P4NM98 A0A2M4BYD1 A0A1S3DTB4 I3MNC2 A0A2M4BYD0 W8BXV1 A0A1U7TG39 A0A1S3JV08 A0A3Q7MPY8 A0A1A8J8X9 A0A1A8D3D3 A0A2U3WT56 H2P2A8 A0A0S2Z4Y5 O60762 A0A2K6V131 A0A384CN11 A0A3Q7WD07 F4WW36 V9L673 A0A3Q2QPG4 A0A2K6EVD4 H3AP13 A0A1A8UCE9 A0A1U7QWB3 A0A3B4B8Y2 I3IZJ1 H0WRY7 M3ZW98 A0A2R9CPW5 H2QKK7
EC Number
2.4.1.83
Pubmed
19121390
26354079
22118469
28756777
18362917
19820115
+ More
20075255 25244985 17204158 17510324 24438588 24845553 26483478 20966253 12364791 29451363 24945155 20920257 23761445 29703783 21993624 25319552 25362486 28004739 16341006 17554307 17975172 26369729 24495485 26871637 9535917 14702039 11780052 15489334 9223280 10835346 17081983 16280320 18691976 20068231 21269460 21406692 22814378 23186163 24275569 25944712 10642597 10642602 15669674 23856421 24813606 21719571 24402279 9215903 25463417 25186727 23542700 22722832 16136131
20075255 25244985 17204158 17510324 24438588 24845553 26483478 20966253 12364791 29451363 24945155 20920257 23761445 29703783 21993624 25319552 25362486 28004739 16341006 17554307 17975172 26369729 24495485 26871637 9535917 14702039 11780052 15489334 9223280 10835346 17081983 16280320 18691976 20068231 21269460 21406692 22814378 23186163 24275569 25944712 10642597 10642602 15669674 23856421 24813606 21719571 24402279 9215903 25463417 25186727 23542700 22722832 16136131
EMBL
DQ443364
ABF51453.1
BABH01040240
BABH01040241
KQ459053
KPJ04203.1
+ More
KQ461181 KPJ07746.1 AGBW02011714 OWR46241.1 NWSH01002285 PCG68678.1 KZ150117 PZC73262.1 ODYU01000663 SOQ35830.1 KQ971354 EFA06224.2 GEDC01006673 JAS30625.1 AAZX01002420 AXCN02002063 AXCM01008671 DQ440282 CH477649 ABF18315.1 EAT37808.1 GECZ01022825 JAS46944.1 KQ435037 KZC14151.1 GANO01001573 JAB58298.1 ATLV01019473 KE525272 KFB44263.1 APCN01001719 KK852423 KDR24245.1 JXUM01025283 KQ560717 KXJ81162.1 CM012443 RVE70278.1 GGFM01002073 MBW22824.1 AAAB01008980 EAA14417.2 GAPW01003838 JAC09760.1 HADW01014757 HADX01002782 SBP16157.1 ADMH02002020 ETN59968.1 NHOQ01000110 PWA32891.1 LNIX01000001 OXA64154.1 HAEB01002872 HAEC01014381 SBQ49399.1 AAKN02050435 KQ759820 OAD62723.1 JU326365 AFE70121.1 AQIA01009143 JV048589 AFI38660.1 GGFK01006142 MBW39463.1 GEZM01081549 JAV61602.1 AAEX03014002 KQ414596 KOC70109.1 GFDL01010019 JAV25026.1 AANG04000928 GDAI01001635 JAI15968.1 CYRY02026032 VCW98605.1 GGFJ01008881 MBW58022.1 AGTP01038186 GGFJ01008882 MBW58023.1 GAMC01000325 JAC06231.1 HAED01019496 HAEE01003284 SBR05961.1 HADZ01021430 HAEA01002389 SBP85371.1 ABGA01105891 ABGA01105892 ABGA01105893 NDHI03003576 PNJ19454.1 KU178444 ALQ33902.1 D86198 D86202 CR456926 AK289569 AL034553 BC007073 BC008466 BC016322 AF007875 GL888404 EGI61625.1 JW874667 AFP07184.1 AFYH01129601 HADY01003686 HAEJ01005323 SBS45780.1 AERX01043460 AAQR03050455 AJFE02050471 AJFE02050472 AJFE02050473 AJFE02050474 AACZ04052869 GABE01011456 NBAG03000253 JAA33283.1 PNI59605.1
KQ461181 KPJ07746.1 AGBW02011714 OWR46241.1 NWSH01002285 PCG68678.1 KZ150117 PZC73262.1 ODYU01000663 SOQ35830.1 KQ971354 EFA06224.2 GEDC01006673 JAS30625.1 AAZX01002420 AXCN02002063 AXCM01008671 DQ440282 CH477649 ABF18315.1 EAT37808.1 GECZ01022825 JAS46944.1 KQ435037 KZC14151.1 GANO01001573 JAB58298.1 ATLV01019473 KE525272 KFB44263.1 APCN01001719 KK852423 KDR24245.1 JXUM01025283 KQ560717 KXJ81162.1 CM012443 RVE70278.1 GGFM01002073 MBW22824.1 AAAB01008980 EAA14417.2 GAPW01003838 JAC09760.1 HADW01014757 HADX01002782 SBP16157.1 ADMH02002020 ETN59968.1 NHOQ01000110 PWA32891.1 LNIX01000001 OXA64154.1 HAEB01002872 HAEC01014381 SBQ49399.1 AAKN02050435 KQ759820 OAD62723.1 JU326365 AFE70121.1 AQIA01009143 JV048589 AFI38660.1 GGFK01006142 MBW39463.1 GEZM01081549 JAV61602.1 AAEX03014002 KQ414596 KOC70109.1 GFDL01010019 JAV25026.1 AANG04000928 GDAI01001635 JAI15968.1 CYRY02026032 VCW98605.1 GGFJ01008881 MBW58022.1 AGTP01038186 GGFJ01008882 MBW58023.1 GAMC01000325 JAC06231.1 HAED01019496 HAEE01003284 SBR05961.1 HADZ01021430 HAEA01002389 SBP85371.1 ABGA01105891 ABGA01105892 ABGA01105893 NDHI03003576 PNJ19454.1 KU178444 ALQ33902.1 D86198 D86202 CR456926 AK289569 AL034553 BC007073 BC008466 BC016322 AF007875 GL888404 EGI61625.1 JW874667 AFP07184.1 AFYH01129601 HADY01003686 HAEJ01005323 SBS45780.1 AERX01043460 AAQR03050455 AJFE02050471 AJFE02050472 AJFE02050473 AJFE02050474 AACZ04052869 GABE01011456 NBAG03000253 JAA33283.1 PNI59605.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000007151
UP000218220
UP000007266
+ More
UP000002358 UP000075886 UP000075885 UP000075900 UP000005203 UP000075883 UP000076408 UP000008820 UP000076502 UP000075884 UP000030765 UP000075840 UP000027135 UP000192223 UP000069940 UP000249989 UP000075882 UP000075881 UP000007062 UP000261560 UP000000673 UP000233080 UP000198287 UP000075903 UP000075902 UP000069272 UP000005447 UP000233140 UP000233120 UP000233100 UP000233180 UP000233200 UP000233060 UP000002254 UP000008225 UP000265040 UP000053825 UP000286640 UP000233020 UP000265180 UP000001038 UP000075880 UP000265200 UP000011712 UP000079169 UP000005215 UP000189704 UP000085678 UP000286641 UP000245340 UP000001595 UP000005640 UP000233220 UP000261680 UP000291021 UP000286642 UP000007755 UP000265000 UP000233160 UP000008672 UP000189706 UP000261520 UP000005207 UP000005225 UP000002852 UP000240080 UP000002277
UP000002358 UP000075886 UP000075885 UP000075900 UP000005203 UP000075883 UP000076408 UP000008820 UP000076502 UP000075884 UP000030765 UP000075840 UP000027135 UP000192223 UP000069940 UP000249989 UP000075882 UP000075881 UP000007062 UP000261560 UP000000673 UP000233080 UP000198287 UP000075903 UP000075902 UP000069272 UP000005447 UP000233140 UP000233120 UP000233100 UP000233180 UP000233200 UP000233060 UP000002254 UP000008225 UP000265040 UP000053825 UP000286640 UP000233020 UP000265180 UP000001038 UP000075880 UP000265200 UP000011712 UP000079169 UP000005215 UP000189704 UP000085678 UP000286641 UP000245340 UP000001595 UP000005640 UP000233220 UP000261680 UP000291021 UP000286642 UP000007755 UP000265000 UP000233160 UP000008672 UP000189706 UP000261520 UP000005207 UP000005225 UP000002852 UP000240080 UP000002277
PRIDE
Pfam
PF00535 Glycos_transf_2
Interpro
SUPFAM
SSF53448
SSF53448
Gene 3D
ProteinModelPortal
Q1HPN8
H9J3X9
A0A194QF83
A0A194QW04
A0A212EXP8
A0A2A4JAG0
+ More
A0A2W1BDH5 A0A2H1V4Q7 D6WPH7 A0A1B6DY92 K7JAA4 A0A182Q9N6 A0A182PFI6 A0A182RHF6 A0A087ZYS0 A0A182M2V4 A0A182YDM0 Q1HR12 A0A1B6FA00 A0A154PQH2 U5EZ22 A0A182NUK2 A0A084W219 A0A182HRL4 A0A067RTV5 A0A1W4X9X5 A0A1W4XKC6 A0A182H4F8 A0A3S2MZF2 A0A182KTP6 A0A2M3Z2T1 A0A182K721 Q7Q1B7 A0A3B3DF72 A0A023EKR8 A0A1A7XDL1 W5J999 A0A2K5IC50 A0A315WAM6 A0A226F3Z6 A0A182UQE5 A0A182U246 A0A182F6A1 A0A1A8EU67 A0A286X9A8 A0A310SWQ5 A0A2K5YCV2 H9F662 A0A2K6CEJ3 A0A2K5W3B5 I0FW34 A0A2K6K3X4 A0A2K6Q8Y9 A0A2K5NX69 A0A2M4AFC4 A0A1Y1KJJ3 J9NZX5 F6RQC7 A0A3Q1H120 A0A0L7RH18 A0A1Q3FBS5 A0A3Q7TYS9 A0A2K5DXH7 A0A3P9LHR4 H2L910 A0A182JKR3 A0A3P9I4S5 M3WC50 A0A0K8TP84 A0A3P4NM98 A0A2M4BYD1 A0A1S3DTB4 I3MNC2 A0A2M4BYD0 W8BXV1 A0A1U7TG39 A0A1S3JV08 A0A3Q7MPY8 A0A1A8J8X9 A0A1A8D3D3 A0A2U3WT56 H2P2A8 A0A0S2Z4Y5 O60762 A0A2K6V131 A0A384CN11 A0A3Q7WD07 F4WW36 V9L673 A0A3Q2QPG4 A0A2K6EVD4 H3AP13 A0A1A8UCE9 A0A1U7QWB3 A0A3B4B8Y2 I3IZJ1 H0WRY7 M3ZW98 A0A2R9CPW5 H2QKK7
A0A2W1BDH5 A0A2H1V4Q7 D6WPH7 A0A1B6DY92 K7JAA4 A0A182Q9N6 A0A182PFI6 A0A182RHF6 A0A087ZYS0 A0A182M2V4 A0A182YDM0 Q1HR12 A0A1B6FA00 A0A154PQH2 U5EZ22 A0A182NUK2 A0A084W219 A0A182HRL4 A0A067RTV5 A0A1W4X9X5 A0A1W4XKC6 A0A182H4F8 A0A3S2MZF2 A0A182KTP6 A0A2M3Z2T1 A0A182K721 Q7Q1B7 A0A3B3DF72 A0A023EKR8 A0A1A7XDL1 W5J999 A0A2K5IC50 A0A315WAM6 A0A226F3Z6 A0A182UQE5 A0A182U246 A0A182F6A1 A0A1A8EU67 A0A286X9A8 A0A310SWQ5 A0A2K5YCV2 H9F662 A0A2K6CEJ3 A0A2K5W3B5 I0FW34 A0A2K6K3X4 A0A2K6Q8Y9 A0A2K5NX69 A0A2M4AFC4 A0A1Y1KJJ3 J9NZX5 F6RQC7 A0A3Q1H120 A0A0L7RH18 A0A1Q3FBS5 A0A3Q7TYS9 A0A2K5DXH7 A0A3P9LHR4 H2L910 A0A182JKR3 A0A3P9I4S5 M3WC50 A0A0K8TP84 A0A3P4NM98 A0A2M4BYD1 A0A1S3DTB4 I3MNC2 A0A2M4BYD0 W8BXV1 A0A1U7TG39 A0A1S3JV08 A0A3Q7MPY8 A0A1A8J8X9 A0A1A8D3D3 A0A2U3WT56 H2P2A8 A0A0S2Z4Y5 O60762 A0A2K6V131 A0A384CN11 A0A3Q7WD07 F4WW36 V9L673 A0A3Q2QPG4 A0A2K6EVD4 H3AP13 A0A1A8UCE9 A0A1U7QWB3 A0A3B4B8Y2 I3IZJ1 H0WRY7 M3ZW98 A0A2R9CPW5 H2QKK7
PDB
5MM1
E-value=5.58814e-33,
Score=350
Ontologies
PATHWAY
GO
PANTHER
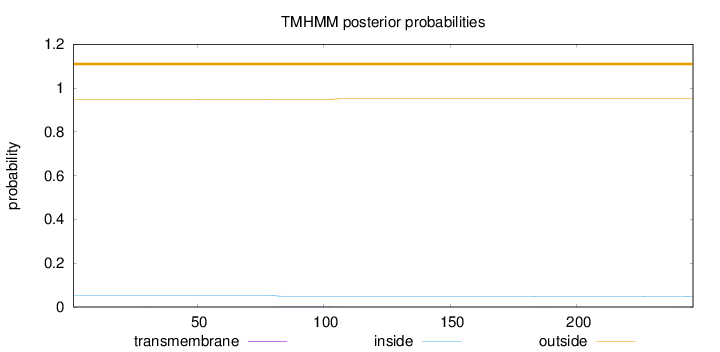
Topology
Subcellular location
Endoplasmic reticulum
Length:
246
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.10063
Exp number, first 60 AAs:
0.00259
Total prob of N-in:
0.05205
outside
1 - 246
Population Genetic Test Statistics
Pi
164.934983
Theta
143.378512
Tajima's D
0.672446
CLR
0.303504
CSRT
0.563421828908555
Interpretation
Uncertain
