Gene
KWMTBOMO12178
Pre Gene Modal
BGIBMGA004220
Annotation
PREDICTED:_heparan-alpha-glucosaminide_N-acetyltransferase-like_[Amyelois_transitella]
Location in the cell
PlasmaMembrane Reliability : 4.79
Sequence
CDS
ATGGGGTTCGGTTTTCAAAATGTCGGCTCATTTATTAGAGAGTGCAATGGTACTACGTTGCGTTACGACCAAGCGTGTTTGAGTATTTTGACCGACGAAGAAGTTACTTTATGGAGCCAATATGCGGAGTGTGATGGGTGCAAATTGCTGCCAACAGCAACGTTCCCGAAGTCTGATCCGATAATAGTCCGCACCTCCGACACTCTGCTCTACGCTCTGAAAAACAGCAGTGGAATAATATGCAAAGGATCCTATCAGTTCGGCGAATATGGACAATACGTTTTCAATCTCACCGAGGGCGCAAAGAATAAATGCCTGCCCCGCATGACTGCGGAACCGGACGCCGCCTACCTCCCGATCCTGACAACGATCCTCATACTCCTGTCCATGGCCACGATGTGGTACGTCGTGAAAGGCATCGGGAAAAGGGTGGTGGCCGGTAGACTATACGCGAGGTTCTTTGCTAGAGAAGATAACGAGCTGGGTTCGGAAACCAGAGTGCTGACCGAGGTGAGCGTGGTGAGGCCCCCGGTGAGGTCCAGGCTGCGGTCACTGGACATCTTCAGGGGATTCGCCATAGCTCTAATGATCTTCGTGAACGCAGGCGGCGGTGGTTACTCGGTGTTCGCTCACAGCACGTGGAATGGGTTAACGGTGGCCGATGTGGTGTTCCCGTGGTTCGCCTTTACCATGGGGGAGGCCCTAGTTCTGTCGCTCAACACTAAGCTTCGCACCGCCATGCCGAGGACTACGGCTTTCTATCAGGTGGTCCGTCGCTCGCTGTTCCTGTCCCTCATCGGGATAGTCCTGGGCTCAGCCGACAGCACCTGGACCGACGTCCGTCTCCCGGGAGTGTTGCAGCGCCTCGCCGCCATGTACCTCATCGTGGGCGCCCTCGAGTGCGCGCTCATGAGGACAACGCAGAACATCACACCAGGCCGTTCATTATTACGGGACATCGCGACCGGGTGGCAGCAGTGGCTCGCTACCATACTGCTCGTTGCTATACAGGTTTGCATATCCCTGACGGTGGCGGCTCCCGGTTGTCCCCGAGGGTACTCCGGTCCAGGTGGGATACATCTCTCGGCCGCCGGCAACCACTCGCTCAAGAACTGTACCGGAGGGATCGCTGGATACATCGATAGAACGCTACTGGGCCCCGCGCATCTGTACCGAGGTGGAACCTTCAAGGAGTTATACCGCACCGTCGTGCCTCACGACCCTGAGGGTATACTGGGCGTCTTCTCAGGAGTGCTGGTGGTGCAGGCTGGCGCTCACGCCACCAGGATCATGCTTGCCTACAACCACGCCAGAGCAAGGATAATGCGCTGGATATTCTGGTCGATAATGTTCGGTGTCGGCGGTGGCGCCCTCTGCATGTTCTCTAAGAACGGCGGCCCGGTGCCCATCAACAAGAACCTGTGGTCGGTGTCCTACTGTCTCGTGACGTCATCGATGGCGTTCTTCATACAGGCCGTGCTGTACTTCATCGTCGATCTGAAGAACAAGTGGGGCGGCAGACCTCTGTATTACGCTGGTCAAAATGCTCTATTCCTTTACGTCGGCTCAGAGCTGTTGAAGCGTCACTTCCCATTGCACTGGTACCTCGAGGCGCCGACGCACGCTCAATTACTGGCAACACACGCCGGCGCCATGTTGATATGGTTAGCGGTCGGCGTTTTCTTGCACAGGAAGAGAATCTTCATAACGTTGTAA
Protein
MGFGFQNVGSFIRECNGTTLRYDQACLSILTDEEVTLWSQYAECDGCKLLPTATFPKSDPIIVRTSDTLLYALKNSSGIICKGSYQFGEYGQYVFNLTEGAKNKCLPRMTAEPDAAYLPILTTILILLSMATMWYVVKGIGKRVVAGRLYARFFAREDNELGSETRVLTEVSVVRPPVRSRLRSLDIFRGFAIALMIFVNAGGGGYSVFAHSTWNGLTVADVVFPWFAFTMGEALVLSLNTKLRTAMPRTTAFYQVVRRSLFLSLIGIVLGSADSTWTDVRLPGVLQRLAAMYLIVGALECALMRTTQNITPGRSLLRDIATGWQQWLATILLVAIQVCISLTVAAPGCPRGYSGPGGIHLSAAGNHSLKNCTGGIAGYIDRTLLGPAHLYRGGTFKELYRTVVPHDPEGILGVFSGVLVVQAGAHATRIMLAYNHARARIMRWIFWSIMFGVGGGALCMFSKNGGPVPINKNLWSVSYCLVTSSMAFFIQAVLYFIVDLKNKWGGRPLYYAGQNALFLYVGSELLKRHFPLHWYLEAPTHAQLLATHAGAMLIWLAVGVFLHRKRIFITL
Summary
Uniprot
A0A2A4JA07
A0A194QL81
H9J3Y1
S4PL18
A0A1E1WPQ5
A0A194QQW9
+ More
A0A1Y1LDE9 A0A2J7RGD1 D6WH02 N6TNX8 U4TT05 A0A023EZN3 A0A0V0G598 A0A1W4XSQ7 A0A087UQB8 A0A0P5E9S8 A0A0P5QQI3 A0A195DMP6 A0A0P4VSJ4 A0A0N8ADU4 A0A0P5C798 A0A0P5N1M7 A0A151WWL7 A0A0P6GFJ3 A0A195CUT6 A0A0P5BL94 A0A026WVR4 A0A3L8DCQ2 A0A232F1H4 T1ITK5 A0A0P6DB80 A0A158NA25 A0A0K2SZ44 A0A0P5SS15 A0A195BKB8 A0A1B6DN10 A0A0P6HGK4 A0A131XSS3 A0A0N8A3Z4 A0A146LU87 A0A0P5LQC3 A0A0P5EAW2 A0A0P5PL02 F4WC35 A0A195FR54 E2BD48 V5I1G4 A0A0L7QQK2 A0A1W4YY52 A0A293LKN0 A0A1S4EG58 A0A0P7VTX6 A0A0L8FIU7 A0A1S3D7K9 F1Q893 A0A1W4YP74 J9JL95 F1NBK1 A0A0Q3PG06 A0A3Q3GZA0 A0A0P5QDD3 A0A0P5ND19 A0A0P4ZCR4 G1SUG4 A0A1L8HW86 A0A0P5HW51 A0A2B4SXF2 A0A3Q1JIG6 A0A3B4TFJ5 G1MZ84 A0A3B5B4C0 A0A087ZR84 A0A2R5LAS3 W4Z2B0 V9IGU7 A0A3Q3KP68 A0A2G8JKG6 A0A1L8E3L4 A0A2K5QNI0 G1NSQ5 A0A3B4YNG0 E9IUZ1 R7VYB6 A0A2A3E690 A0A091KJ11
A0A1Y1LDE9 A0A2J7RGD1 D6WH02 N6TNX8 U4TT05 A0A023EZN3 A0A0V0G598 A0A1W4XSQ7 A0A087UQB8 A0A0P5E9S8 A0A0P5QQI3 A0A195DMP6 A0A0P4VSJ4 A0A0N8ADU4 A0A0P5C798 A0A0P5N1M7 A0A151WWL7 A0A0P6GFJ3 A0A195CUT6 A0A0P5BL94 A0A026WVR4 A0A3L8DCQ2 A0A232F1H4 T1ITK5 A0A0P6DB80 A0A158NA25 A0A0K2SZ44 A0A0P5SS15 A0A195BKB8 A0A1B6DN10 A0A0P6HGK4 A0A131XSS3 A0A0N8A3Z4 A0A146LU87 A0A0P5LQC3 A0A0P5EAW2 A0A0P5PL02 F4WC35 A0A195FR54 E2BD48 V5I1G4 A0A0L7QQK2 A0A1W4YY52 A0A293LKN0 A0A1S4EG58 A0A0P7VTX6 A0A0L8FIU7 A0A1S3D7K9 F1Q893 A0A1W4YP74 J9JL95 F1NBK1 A0A0Q3PG06 A0A3Q3GZA0 A0A0P5QDD3 A0A0P5ND19 A0A0P4ZCR4 G1SUG4 A0A1L8HW86 A0A0P5HW51 A0A2B4SXF2 A0A3Q1JIG6 A0A3B4TFJ5 G1MZ84 A0A3B5B4C0 A0A087ZR84 A0A2R5LAS3 W4Z2B0 V9IGU7 A0A3Q3KP68 A0A2G8JKG6 A0A1L8E3L4 A0A2K5QNI0 G1NSQ5 A0A3B4YNG0 E9IUZ1 R7VYB6 A0A2A3E690 A0A091KJ11
Pubmed
EMBL
NWSH01002285
PCG68679.1
KQ459053
KPJ04201.1
BABH01040235
BABH01040236
+ More
BABH01040237 BABH01040238 GAIX01004260 JAA88300.1 GDQN01002074 JAT88980.1 KQ461181 KPJ07744.1 GEZM01062601 JAV69596.1 NEVH01004401 PNF39887.1 KQ971321 EFA00134.1 APGK01026887 KB740648 ENN79718.1 KB630779 KB631357 ERL83862.1 ERL84257.1 GBBI01003990 JAC14722.1 GECL01002904 JAP03220.1 KK121017 KFM79557.1 GDIP01146179 GDIQ01097084 LRGB01000024 JAJ77223.1 JAL54642.1 KZS21384.1 GDIQ01131289 JAL20437.1 KQ980734 KYN13774.1 GDKW01001671 JAI54924.1 GDIP01157185 JAJ66217.1 GDIP01174581 JAJ48821.1 GDIQ01148681 JAL03045.1 KQ982686 KYQ52304.1 GDIQ01036576 JAN58161.1 KQ977276 KYN04446.1 GDIP01187091 JAJ36311.1 KK107079 EZA60115.1 QOIP01000010 RLU17659.1 NNAY01001324 OXU24360.1 JH431487 GDIQ01088487 JAN06250.1 ADTU01009953 HACA01001657 CDW19018.1 GDIP01136126 JAL67588.1 KQ976455 KYM85093.1 GEDC01010212 JAS27086.1 GDIQ01019133 JAN75604.1 GEFM01006518 JAP69278.1 GDIP01184785 JAJ38617.1 GDHC01008327 GDHC01008187 JAQ10302.1 JAQ10442.1 GDIQ01166724 JAK85001.1 GDIP01144175 JAJ79227.1 GDIQ01127428 JAL24298.1 GL888070 EGI68015.1 KQ981305 KYN42911.1 GL447563 EFN86390.1 GANP01001361 JAB83107.1 KQ414786 KOC60900.1 GFWV01009318 MAA34047.1 JARO02000718 KPP77624.1 KQ430684 KOF63776.1 BX511256 ABLF02037236 ABLF02037244 AADN05000017 LMAW01002573 KQK79683.1 GDIQ01116237 JAL35489.1 GDIQ01147132 JAL04594.1 GDIP01214317 JAJ09085.1 CM004466 OCU00375.1 GDIQ01227992 JAK23733.1 LSMT01000008 PFX33869.1 GGLE01002456 MBY06582.1 AAGJ04099310 AAGJ04099311 AAGJ04099312 AAGJ04099313 JR046973 AEY60290.1 MRZV01001714 PIK36227.1 GFDF01000922 JAV13162.1 AAPE02033681 GL766098 EFZ15612.1 KB375379 EMC90042.1 KZ288375 PBC26689.1 KK530449 KFP27834.1
BABH01040237 BABH01040238 GAIX01004260 JAA88300.1 GDQN01002074 JAT88980.1 KQ461181 KPJ07744.1 GEZM01062601 JAV69596.1 NEVH01004401 PNF39887.1 KQ971321 EFA00134.1 APGK01026887 KB740648 ENN79718.1 KB630779 KB631357 ERL83862.1 ERL84257.1 GBBI01003990 JAC14722.1 GECL01002904 JAP03220.1 KK121017 KFM79557.1 GDIP01146179 GDIQ01097084 LRGB01000024 JAJ77223.1 JAL54642.1 KZS21384.1 GDIQ01131289 JAL20437.1 KQ980734 KYN13774.1 GDKW01001671 JAI54924.1 GDIP01157185 JAJ66217.1 GDIP01174581 JAJ48821.1 GDIQ01148681 JAL03045.1 KQ982686 KYQ52304.1 GDIQ01036576 JAN58161.1 KQ977276 KYN04446.1 GDIP01187091 JAJ36311.1 KK107079 EZA60115.1 QOIP01000010 RLU17659.1 NNAY01001324 OXU24360.1 JH431487 GDIQ01088487 JAN06250.1 ADTU01009953 HACA01001657 CDW19018.1 GDIP01136126 JAL67588.1 KQ976455 KYM85093.1 GEDC01010212 JAS27086.1 GDIQ01019133 JAN75604.1 GEFM01006518 JAP69278.1 GDIP01184785 JAJ38617.1 GDHC01008327 GDHC01008187 JAQ10302.1 JAQ10442.1 GDIQ01166724 JAK85001.1 GDIP01144175 JAJ79227.1 GDIQ01127428 JAL24298.1 GL888070 EGI68015.1 KQ981305 KYN42911.1 GL447563 EFN86390.1 GANP01001361 JAB83107.1 KQ414786 KOC60900.1 GFWV01009318 MAA34047.1 JARO02000718 KPP77624.1 KQ430684 KOF63776.1 BX511256 ABLF02037236 ABLF02037244 AADN05000017 LMAW01002573 KQK79683.1 GDIQ01116237 JAL35489.1 GDIQ01147132 JAL04594.1 GDIP01214317 JAJ09085.1 CM004466 OCU00375.1 GDIQ01227992 JAK23733.1 LSMT01000008 PFX33869.1 GGLE01002456 MBY06582.1 AAGJ04099310 AAGJ04099311 AAGJ04099312 AAGJ04099313 JR046973 AEY60290.1 MRZV01001714 PIK36227.1 GFDF01000922 JAV13162.1 AAPE02033681 GL766098 EFZ15612.1 KB375379 EMC90042.1 KZ288375 PBC26689.1 KK530449 KFP27834.1
Proteomes
UP000218220
UP000053268
UP000005204
UP000053240
UP000235965
UP000007266
+ More
UP000019118 UP000030742 UP000192223 UP000054359 UP000076858 UP000078492 UP000075809 UP000078542 UP000053097 UP000279307 UP000215335 UP000005205 UP000078540 UP000007755 UP000078541 UP000008237 UP000053825 UP000192224 UP000079169 UP000034805 UP000053454 UP000000437 UP000007819 UP000000539 UP000051836 UP000261660 UP000001811 UP000186698 UP000225706 UP000265040 UP000261420 UP000001645 UP000261400 UP000005203 UP000007110 UP000261640 UP000230750 UP000233040 UP000001074 UP000261360 UP000242457
UP000019118 UP000030742 UP000192223 UP000054359 UP000076858 UP000078492 UP000075809 UP000078542 UP000053097 UP000279307 UP000215335 UP000005205 UP000078540 UP000007755 UP000078541 UP000008237 UP000053825 UP000192224 UP000079169 UP000034805 UP000053454 UP000000437 UP000007819 UP000000539 UP000051836 UP000261660 UP000001811 UP000186698 UP000225706 UP000265040 UP000261420 UP000001645 UP000261400 UP000005203 UP000007110 UP000261640 UP000230750 UP000233040 UP000001074 UP000261360 UP000242457
PRIDE
ProteinModelPortal
A0A2A4JA07
A0A194QL81
H9J3Y1
S4PL18
A0A1E1WPQ5
A0A194QQW9
+ More
A0A1Y1LDE9 A0A2J7RGD1 D6WH02 N6TNX8 U4TT05 A0A023EZN3 A0A0V0G598 A0A1W4XSQ7 A0A087UQB8 A0A0P5E9S8 A0A0P5QQI3 A0A195DMP6 A0A0P4VSJ4 A0A0N8ADU4 A0A0P5C798 A0A0P5N1M7 A0A151WWL7 A0A0P6GFJ3 A0A195CUT6 A0A0P5BL94 A0A026WVR4 A0A3L8DCQ2 A0A232F1H4 T1ITK5 A0A0P6DB80 A0A158NA25 A0A0K2SZ44 A0A0P5SS15 A0A195BKB8 A0A1B6DN10 A0A0P6HGK4 A0A131XSS3 A0A0N8A3Z4 A0A146LU87 A0A0P5LQC3 A0A0P5EAW2 A0A0P5PL02 F4WC35 A0A195FR54 E2BD48 V5I1G4 A0A0L7QQK2 A0A1W4YY52 A0A293LKN0 A0A1S4EG58 A0A0P7VTX6 A0A0L8FIU7 A0A1S3D7K9 F1Q893 A0A1W4YP74 J9JL95 F1NBK1 A0A0Q3PG06 A0A3Q3GZA0 A0A0P5QDD3 A0A0P5ND19 A0A0P4ZCR4 G1SUG4 A0A1L8HW86 A0A0P5HW51 A0A2B4SXF2 A0A3Q1JIG6 A0A3B4TFJ5 G1MZ84 A0A3B5B4C0 A0A087ZR84 A0A2R5LAS3 W4Z2B0 V9IGU7 A0A3Q3KP68 A0A2G8JKG6 A0A1L8E3L4 A0A2K5QNI0 G1NSQ5 A0A3B4YNG0 E9IUZ1 R7VYB6 A0A2A3E690 A0A091KJ11
A0A1Y1LDE9 A0A2J7RGD1 D6WH02 N6TNX8 U4TT05 A0A023EZN3 A0A0V0G598 A0A1W4XSQ7 A0A087UQB8 A0A0P5E9S8 A0A0P5QQI3 A0A195DMP6 A0A0P4VSJ4 A0A0N8ADU4 A0A0P5C798 A0A0P5N1M7 A0A151WWL7 A0A0P6GFJ3 A0A195CUT6 A0A0P5BL94 A0A026WVR4 A0A3L8DCQ2 A0A232F1H4 T1ITK5 A0A0P6DB80 A0A158NA25 A0A0K2SZ44 A0A0P5SS15 A0A195BKB8 A0A1B6DN10 A0A0P6HGK4 A0A131XSS3 A0A0N8A3Z4 A0A146LU87 A0A0P5LQC3 A0A0P5EAW2 A0A0P5PL02 F4WC35 A0A195FR54 E2BD48 V5I1G4 A0A0L7QQK2 A0A1W4YY52 A0A293LKN0 A0A1S4EG58 A0A0P7VTX6 A0A0L8FIU7 A0A1S3D7K9 F1Q893 A0A1W4YP74 J9JL95 F1NBK1 A0A0Q3PG06 A0A3Q3GZA0 A0A0P5QDD3 A0A0P5ND19 A0A0P4ZCR4 G1SUG4 A0A1L8HW86 A0A0P5HW51 A0A2B4SXF2 A0A3Q1JIG6 A0A3B4TFJ5 G1MZ84 A0A3B5B4C0 A0A087ZR84 A0A2R5LAS3 W4Z2B0 V9IGU7 A0A3Q3KP68 A0A2G8JKG6 A0A1L8E3L4 A0A2K5QNI0 G1NSQ5 A0A3B4YNG0 E9IUZ1 R7VYB6 A0A2A3E690 A0A091KJ11
Ontologies
PATHWAY
PANTHER
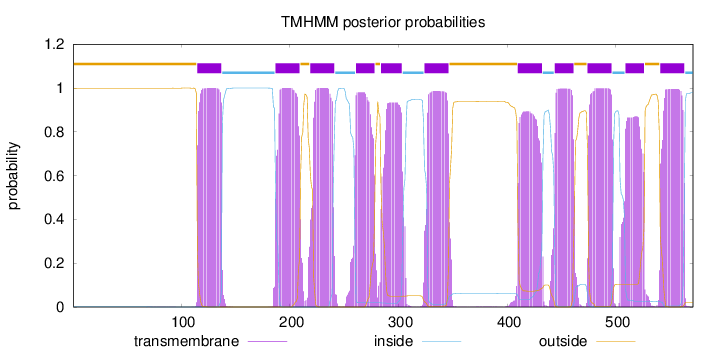
Topology
Length:
571
Number of predicted TMHs:
11
Exp number of AAs in TMHs:
225.49167
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00108
outside
1 - 114
TMhelix
115 - 137
inside
138 - 186
TMhelix
187 - 209
outside
210 - 218
TMhelix
219 - 241
inside
242 - 260
TMhelix
261 - 278
outside
279 - 283
TMhelix
284 - 303
inside
304 - 323
TMhelix
324 - 346
outside
347 - 409
TMhelix
410 - 432
inside
433 - 443
TMhelix
444 - 461
outside
462 - 473
TMhelix
474 - 496
inside
497 - 508
TMhelix
509 - 526
outside
527 - 540
TMhelix
541 - 563
inside
564 - 571
Population Genetic Test Statistics
Pi
194.586643
Theta
218.718138
Tajima's D
0.513746
CLR
0.010045
CSRT
0.513224338783061
Interpretation
Uncertain
