Gene
KWMTBOMO12177 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA004221
Annotation
glucose-6-phosphate_isomerase_[Bombyx_mori]
Full name
Glucose-6-phosphate isomerase
Location in the cell
Cytoplasmic Reliability : 3.639
Sequence
CDS
ATGGAACCGAAAATTAATTTAAAGCAAGACGCAGCTTATCAAAAATTACAAGAATATTACAATGTAAACAGCACAAAAATTAATATGCTCCAACTTTTCCAACAAGATCGGGAGCGTTTCGAAAAGTTCAGCCTCTGTATCCCAACACCGAACGATGGAGACATCCTACTTGACTATTCCAAGAACCGTATCAACAGTGATGTTTTTAAGTTGCTCCTCGATCTCGCTAAGAGCAGAAATGTTGAACAAGCCAGAGATGCCATGTTCGCAGGTCAAAAGATAAACTTCACAGAAGACAGGGCAGTACTGCATATAGCTCTGCGTAACCGTCAGAACAAGCCTATCCTGGTGAACGGTAAGGATGTCAGCACTGATGTCAATGCGGTACTAGAACACATGAAGGAATTCTCCGATCAAGTAGTAAGCGGGCAATGGAAGGGGTATACGGGAAAAGCTATAACAGACGTGATCAACATTGGTATTGGAGGCTCAGATGTGGGTCCTCTCATGGTCACAGAAGCACTGAAGCCCTATGCTAATCATCTTAAGGTCCATTTCGTATCTAACATCGACGGCACCCACCTGGCTGAGGTCTTGAAGAAGCTGAACCCTGAGACAGCGCTGTTCATCATAGCATCCAAGACTTTCACCACACAAGAGACGATAACCAATGCGAGCTCAGCTAAGACGTGGTTCCTGGAATCCGCTAAGGATCCCTCGGCGGTGGCGAGACACTTCGTCGCTTTATCGACGAACGCCGAGAAGGTCACCGCGTTCGGTATCGACGCTAACAACATGTTCGGCTTCTGGGACTGGGTCGGGGGCAGATATTCGCTGTGGTCCGCGATCGGTCTGTCCATCTCGCTGTACATCGGACACGAAAACTTCGAGAAACTTCTGGACGGAGCCAATTTCATGGACCAGCACTTCGTCACCGCACCGCTGGAGAAGAATGCTCCCGTGATCTTAGCTCTGTTAGGAGTCTGGTACCACAATTTCTACGGAGCAGAGACCCATGCGCTGCTGCCTTACGATCAATATCTACACAGATTCGCAGCGTATTTCCAGCAGGGCGATATGGAGAGCAATGGCAAGTACGTGACGCGTTCAGGAGCCGAAGTGGAATACTCCACGGGGCCCATCGTGTGGGGCGAGCCCGGGACCAACGGACAGCACGCCTTCTACCAGCTCATACACCAGGGAACCAGATTGATTCCATGCGATTTCATCGCTCCAGCCCAAACTCACAATCCAATTTCGAATGGTGTTCACCACAAGATCCTTCTAGCTAATTTCTTGGCTCAAACCGAAGCCCTGATGAAGGGCAAGACAGCTGACGAGGCTAAAGCTGAGCTAGAGAAATCTGGTATGGCTCCGGAGGCAATAGACAAGATCCTTCCGCACAAGGTATTCAAAGGAAACCGGCCTACAAACTCTATTGTTCTGAAGAAGATCACTCCATTCACTTTAGGAGCACTTATTGCCATGTACGAACACAAGATCTTCACGCAAGGTGTGATCTGGGATATCAATTCATACGATCAATGGGGAGTTGAACTCGGCAAGCAATTGGCCAAGGCAATCGAGCCTGAGCTGCAGGGGACTGCAGCTGTGACCGGCCACGACGCTTCCACGAATGGACTCATTAACTTCCTCAAGAAGAACTTTTGA
Protein
MEPKINLKQDAAYQKLQEYYNVNSTKINMLQLFQQDRERFEKFSLCIPTPNDGDILLDYSKNRINSDVFKLLLDLAKSRNVEQARDAMFAGQKINFTEDRAVLHIALRNRQNKPILVNGKDVSTDVNAVLEHMKEFSDQVVSGQWKGYTGKAITDVINIGIGGSDVGPLMVTEALKPYANHLKVHFVSNIDGTHLAEVLKKLNPETALFIIASKTFTTQETITNASSAKTWFLESAKDPSAVARHFVALSTNAEKVTAFGIDANNMFGFWDWVGGRYSLWSAIGLSISLYIGHENFEKLLDGANFMDQHFVTAPLEKNAPVILALLGVWYHNFYGAETHALLPYDQYLHRFAAYFQQGDMESNGKYVTRSGAEVEYSTGPIVWGEPGTNGQHAFYQLIHQGTRLIPCDFIAPAQTHNPISNGVHHKILLANFLAQTEALMKGKTADEAKAELEKSGMAPEAIDKILPHKVFKGNRPTNSIVLKKITPFTLGALIAMYEHKIFTQGVIWDINSYDQWGVELGKQLAKAIEPELQGTAAVTGHDASTNGLINFLKKNF
Summary
Catalytic Activity
aldehydo-D-glucose 6-phosphate = keto-D-fructose 6-phosphate
Similarity
Belongs to the GPI family.
Uniprot
Q2F5I6
H9J3Y2
Q2Q4Z1
C5I8Y2
C5I905
G9BZI8
+ More
G9BZI3 C5I8Y4 Q2Q4Y6 G9BZI5 Q2Q4Z5 Q2F7Z2 C5I8Z2 A0A2H1V8Q8 C5I8X3 Q2Q4Z7 C5I8X0 C5I904 C5I902 C5I8X1 Q2Q4Z6 C5I900 C5I903 C5I8Z9 C5I8W3 C5I8X9 C5I8X8 C5I8Y5 C5I906 G9BZI7 Q2Q4Z9 C5I907 C5I8X4 G9BZJ0 C5I8X7 C5I8W2 C5I8X6 C5I8Y1 C5I8W5 H6TMR9 C5I8W6 C5I8Y3 C5I8W7 C5I8W9 H6TMR8 Q2Q4Y7 Q2Q4Z8 G9BZJ1 C5I8X5 Q2Q4Y8 Q2Q4Z0 C5I8Y0 C5I8W4 C5I8Z5 C5I8W8 G9BZI2 C5I901 C5I8Z3 G9BZI6 E3UKM3 G9BZI9 H6TMT1 C5I8Z4 C5I8Z8 C5I8Z7 H6TMQ3 C5I8Y8 D6BNT9 H6TMQ8 H6TMS5 H6TMQ6 H6TMR5 H6TMT2 C5I8Z0 H6TMR6 H6TMS2 H6TMR0 H6TMQ9 Q2Q4Z2 H6TMQ5 H6TMP9 Q2Q4Y9 H6TMS4 D2XVJ3 C5I8Y9 C5I8Y6 H6TMT0 C6FF88 D2XVG0 H6TMT5 H6TMR3 C6FFA4 C6FFA5 D2XVI5 D2XVJ1 H6TMQ0 H6TMR4 D2XVJ0 H6TMR1 H6TMQ7
G9BZI3 C5I8Y4 Q2Q4Y6 G9BZI5 Q2Q4Z5 Q2F7Z2 C5I8Z2 A0A2H1V8Q8 C5I8X3 Q2Q4Z7 C5I8X0 C5I904 C5I902 C5I8X1 Q2Q4Z6 C5I900 C5I903 C5I8Z9 C5I8W3 C5I8X9 C5I8X8 C5I8Y5 C5I906 G9BZI7 Q2Q4Z9 C5I907 C5I8X4 G9BZJ0 C5I8X7 C5I8W2 C5I8X6 C5I8Y1 C5I8W5 H6TMR9 C5I8W6 C5I8Y3 C5I8W7 C5I8W9 H6TMR8 Q2Q4Y7 Q2Q4Z8 G9BZJ1 C5I8X5 Q2Q4Y8 Q2Q4Z0 C5I8Y0 C5I8W4 C5I8Z5 C5I8W8 G9BZI2 C5I901 C5I8Z3 G9BZI6 E3UKM3 G9BZI9 H6TMT1 C5I8Z4 C5I8Z8 C5I8Z7 H6TMQ3 C5I8Y8 D6BNT9 H6TMQ8 H6TMS5 H6TMQ6 H6TMR5 H6TMT2 C5I8Z0 H6TMR6 H6TMS2 H6TMR0 H6TMQ9 Q2Q4Z2 H6TMQ5 H6TMP9 Q2Q4Y9 H6TMS4 D2XVJ3 C5I8Y9 C5I8Y6 H6TMT0 C6FF88 D2XVG0 H6TMT5 H6TMR3 C6FFA4 C6FFA5 D2XVI5 D2XVJ1 H6TMQ0 H6TMR4 D2XVJ0 H6TMR1 H6TMQ7
EC Number
5.3.1.9
EMBL
DQ311437
ABD36381.1
BABH01040234
BABH01040235
DQ205071
FJ796737
+ More
ABB76196.1 ACS27520.1 FJ796728 ACS27511.1 FJ796751 ACS27534.1 HQ717714 AEV89434.1 HQ717709 AEV89429.1 FJ796730 ACS27513.1 DQ205076 ABB76201.1 HQ717711 AEV89431.1 DQ205067 ABB76192.1 DQ205092 ABB00833.1 FJ796738 ACS27521.1 ODYU01001261 SOQ37230.1 FJ796719 ACS27502.1 DQ205065 FJ796718 ABB76190.1 ACS27501.1 FJ796716 ACS27499.1 FJ796750 ACS27533.1 FJ796748 ACS27531.1 FJ796717 ACS27500.1 DQ205066 ABB76191.1 FJ796746 ACS27529.1 FJ796749 ACS27532.1 FJ796745 ACS27528.1 FJ796709 ACS27492.1 FJ796725 HQ717710 ACS27508.1 AEV89430.1 FJ796724 ACS27507.1 FJ796731 ACS27514.1 FJ796752 ACS27535.1 HQ717713 AEV89433.1 DQ205063 ABB76188.1 FJ796753 ACS27536.1 FJ796720 ACS27503.1 HQ717716 AEV89436.1 FJ796723 ACS27506.1 FJ796708 ACS27491.1 FJ796722 ACS27505.1 FJ796727 ACS27510.1 FJ796711 ACS27494.1 JN191731 AFA51145.1 FJ796712 ACS27495.1 FJ796729 ACS27512.1 FJ796713 ACS27496.1 FJ796715 ACS27498.1 JN191730 AFA51144.1 DQ205075 ABB76200.1 DQ205064 ABB76189.1 HQ717717 AEV89437.1 FJ796721 ACS27504.1 DQ205074 ABB76199.1 DQ205072 ABB76197.1 FJ796726 ACS27509.1 FJ796710 ACS27493.1 FJ796741 FJ796742 ACS27524.1 FJ796714 ACS27497.1 HQ717708 AEV89428.1 FJ796747 ACS27530.1 FJ796739 ACS27522.1 HQ717712 AEV89432.1 HM449897 ADO33032.1 HQ717715 AEV89435.1 JN191743 AFA51157.1 FJ796740 ACS27523.1 FJ796744 ACS27527.1 FJ796743 ACS27526.1 JN191715 JN191716 AFA51129.1 FJ796734 ACS27517.1 FJ793040 ACV97159.1 JN191720 AFA51134.1 JN191737 AFA51151.1 JN191718 AFA51132.1 JN191727 AFA51141.1 JN191744 JN191745 JN191746 AFA51158.1 FJ796736 ACS27519.1 JN191728 AFA51142.1 JN191734 AFA51148.1 JN191722 AFA51136.1 JN191721 AFA51135.1 DQ205070 ABB76195.1 JN191717 AFA51131.1 JN191711 AFA51125.1 DQ205073 ABB76198.1 JN191736 AFA51150.1 GU263977 ADB11225.1 FJ796735 ACS27518.1 FJ796732 ACS27515.1 JN191742 AFA51156.1 EU888447 ACF57681.1 GU263944 ADB11192.1 JN191747 AFA51161.1 JN191725 AFA51139.1 EU888463 EU888465 EU888470 GU263964 GU263965 GU263966 GU263967 GU263968 GU263970 GU263971 GU263972 GU263973 GU263976 ACF57697.1 ADB11212.1 ADB11214.1 ADB11215.1 ADB11216.1 ADB11218.1 ADB11219.1 ADB11220.1 ADB11224.1 EU888464 GU263937 GU263938 GU263939 GU263940 GU263941 ACF57698.1 ADB11185.1 ADB11187.1 ADB11188.1 ADB11189.1 GU263969 ADB11217.1 GU263975 ADB11223.1 JN191712 AFA51126.1 JN191726 AFA51140.1 GU263974 ADB11222.1 JN191723 AFA51137.1 JN191719 AFA51133.1
ABB76196.1 ACS27520.1 FJ796728 ACS27511.1 FJ796751 ACS27534.1 HQ717714 AEV89434.1 HQ717709 AEV89429.1 FJ796730 ACS27513.1 DQ205076 ABB76201.1 HQ717711 AEV89431.1 DQ205067 ABB76192.1 DQ205092 ABB00833.1 FJ796738 ACS27521.1 ODYU01001261 SOQ37230.1 FJ796719 ACS27502.1 DQ205065 FJ796718 ABB76190.1 ACS27501.1 FJ796716 ACS27499.1 FJ796750 ACS27533.1 FJ796748 ACS27531.1 FJ796717 ACS27500.1 DQ205066 ABB76191.1 FJ796746 ACS27529.1 FJ796749 ACS27532.1 FJ796745 ACS27528.1 FJ796709 ACS27492.1 FJ796725 HQ717710 ACS27508.1 AEV89430.1 FJ796724 ACS27507.1 FJ796731 ACS27514.1 FJ796752 ACS27535.1 HQ717713 AEV89433.1 DQ205063 ABB76188.1 FJ796753 ACS27536.1 FJ796720 ACS27503.1 HQ717716 AEV89436.1 FJ796723 ACS27506.1 FJ796708 ACS27491.1 FJ796722 ACS27505.1 FJ796727 ACS27510.1 FJ796711 ACS27494.1 JN191731 AFA51145.1 FJ796712 ACS27495.1 FJ796729 ACS27512.1 FJ796713 ACS27496.1 FJ796715 ACS27498.1 JN191730 AFA51144.1 DQ205075 ABB76200.1 DQ205064 ABB76189.1 HQ717717 AEV89437.1 FJ796721 ACS27504.1 DQ205074 ABB76199.1 DQ205072 ABB76197.1 FJ796726 ACS27509.1 FJ796710 ACS27493.1 FJ796741 FJ796742 ACS27524.1 FJ796714 ACS27497.1 HQ717708 AEV89428.1 FJ796747 ACS27530.1 FJ796739 ACS27522.1 HQ717712 AEV89432.1 HM449897 ADO33032.1 HQ717715 AEV89435.1 JN191743 AFA51157.1 FJ796740 ACS27523.1 FJ796744 ACS27527.1 FJ796743 ACS27526.1 JN191715 JN191716 AFA51129.1 FJ796734 ACS27517.1 FJ793040 ACV97159.1 JN191720 AFA51134.1 JN191737 AFA51151.1 JN191718 AFA51132.1 JN191727 AFA51141.1 JN191744 JN191745 JN191746 AFA51158.1 FJ796736 ACS27519.1 JN191728 AFA51142.1 JN191734 AFA51148.1 JN191722 AFA51136.1 JN191721 AFA51135.1 DQ205070 ABB76195.1 JN191717 AFA51131.1 JN191711 AFA51125.1 DQ205073 ABB76198.1 JN191736 AFA51150.1 GU263977 ADB11225.1 FJ796735 ACS27518.1 FJ796732 ACS27515.1 JN191742 AFA51156.1 EU888447 ACF57681.1 GU263944 ADB11192.1 JN191747 AFA51161.1 JN191725 AFA51139.1 EU888463 EU888465 EU888470 GU263964 GU263965 GU263966 GU263967 GU263968 GU263970 GU263971 GU263972 GU263973 GU263976 ACF57697.1 ADB11212.1 ADB11214.1 ADB11215.1 ADB11216.1 ADB11218.1 ADB11219.1 ADB11220.1 ADB11224.1 EU888464 GU263937 GU263938 GU263939 GU263940 GU263941 ACF57698.1 ADB11185.1 ADB11187.1 ADB11188.1 ADB11189.1 GU263969 ADB11217.1 GU263975 ADB11223.1 JN191712 AFA51126.1 JN191726 AFA51140.1 GU263974 ADB11222.1 JN191723 AFA51137.1 JN191719 AFA51133.1
Proteomes
Pfam
PF00342 PGI
Interpro
Gene 3D
ProteinModelPortal
Q2F5I6
H9J3Y2
Q2Q4Z1
C5I8Y2
C5I905
G9BZI8
+ More
G9BZI3 C5I8Y4 Q2Q4Y6 G9BZI5 Q2Q4Z5 Q2F7Z2 C5I8Z2 A0A2H1V8Q8 C5I8X3 Q2Q4Z7 C5I8X0 C5I904 C5I902 C5I8X1 Q2Q4Z6 C5I900 C5I903 C5I8Z9 C5I8W3 C5I8X9 C5I8X8 C5I8Y5 C5I906 G9BZI7 Q2Q4Z9 C5I907 C5I8X4 G9BZJ0 C5I8X7 C5I8W2 C5I8X6 C5I8Y1 C5I8W5 H6TMR9 C5I8W6 C5I8Y3 C5I8W7 C5I8W9 H6TMR8 Q2Q4Y7 Q2Q4Z8 G9BZJ1 C5I8X5 Q2Q4Y8 Q2Q4Z0 C5I8Y0 C5I8W4 C5I8Z5 C5I8W8 G9BZI2 C5I901 C5I8Z3 G9BZI6 E3UKM3 G9BZI9 H6TMT1 C5I8Z4 C5I8Z8 C5I8Z7 H6TMQ3 C5I8Y8 D6BNT9 H6TMQ8 H6TMS5 H6TMQ6 H6TMR5 H6TMT2 C5I8Z0 H6TMR6 H6TMS2 H6TMR0 H6TMQ9 Q2Q4Z2 H6TMQ5 H6TMP9 Q2Q4Y9 H6TMS4 D2XVJ3 C5I8Y9 C5I8Y6 H6TMT0 C6FF88 D2XVG0 H6TMT5 H6TMR3 C6FFA4 C6FFA5 D2XVI5 D2XVJ1 H6TMQ0 H6TMR4 D2XVJ0 H6TMR1 H6TMQ7
G9BZI3 C5I8Y4 Q2Q4Y6 G9BZI5 Q2Q4Z5 Q2F7Z2 C5I8Z2 A0A2H1V8Q8 C5I8X3 Q2Q4Z7 C5I8X0 C5I904 C5I902 C5I8X1 Q2Q4Z6 C5I900 C5I903 C5I8Z9 C5I8W3 C5I8X9 C5I8X8 C5I8Y5 C5I906 G9BZI7 Q2Q4Z9 C5I907 C5I8X4 G9BZJ0 C5I8X7 C5I8W2 C5I8X6 C5I8Y1 C5I8W5 H6TMR9 C5I8W6 C5I8Y3 C5I8W7 C5I8W9 H6TMR8 Q2Q4Y7 Q2Q4Z8 G9BZJ1 C5I8X5 Q2Q4Y8 Q2Q4Z0 C5I8Y0 C5I8W4 C5I8Z5 C5I8W8 G9BZI2 C5I901 C5I8Z3 G9BZI6 E3UKM3 G9BZI9 H6TMT1 C5I8Z4 C5I8Z8 C5I8Z7 H6TMQ3 C5I8Y8 D6BNT9 H6TMQ8 H6TMS5 H6TMQ6 H6TMR5 H6TMT2 C5I8Z0 H6TMR6 H6TMS2 H6TMR0 H6TMQ9 Q2Q4Z2 H6TMQ5 H6TMP9 Q2Q4Y9 H6TMS4 D2XVJ3 C5I8Y9 C5I8Y6 H6TMT0 C6FF88 D2XVG0 H6TMT5 H6TMR3 C6FFA4 C6FFA5 D2XVI5 D2XVJ1 H6TMQ0 H6TMR4 D2XVJ0 H6TMR1 H6TMQ7
PDB
4WMJ
E-value=0,
Score=2649
Ontologies
KEGG
PATHWAY
00010
Glycolysis / Gluconeogenesis - Bombyx mori (domestic silkworm)
00030 Pentose phosphate pathway - Bombyx mori (domestic silkworm)
00500 Starch and sucrose metabolism - Bombyx mori (domestic silkworm)
00520 Amino sugar and nucleotide sugar metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
01200 Carbon metabolism - Bombyx mori (domestic silkworm)
00030 Pentose phosphate pathway - Bombyx mori (domestic silkworm)
00500 Starch and sucrose metabolism - Bombyx mori (domestic silkworm)
00520 Amino sugar and nucleotide sugar metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
01200 Carbon metabolism - Bombyx mori (domestic silkworm)
GO
PANTHER
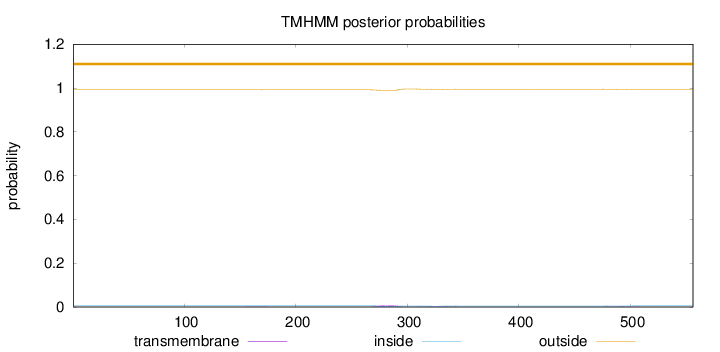
Topology
Length:
556
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.23281
Exp number, first 60 AAs:
0.00025
Total prob of N-in:
0.00816
outside
1 - 556
Population Genetic Test Statistics
Pi
180.207019
Theta
174.484531
Tajima's D
0.989326
CLR
0.21849
CSRT
0.661366931653417
Interpretation
Uncertain
