Gene
KWMTBOMO12175
Annotation
PREDICTED:_plasminogen_receptor_(KT)_[Amyelois_transitella]
Location in the cell
Mitochondrial Reliability : 1.024 Nuclear Reliability : 1.583
Sequence
CDS
ATGGGTAATTTGTTCAGCGTCAGCATGGAAGAAAATTACAAAAGGAACCAAGAATTCTTGCAAACTATAAACAAAGTCAAGATGGAGAGACAAATACAACTACAATATCAAATGCAAGAACGCCAAATGGCCTTGGAAATAGCTAAAAACAGAGACACATGTCTGTGGATTACTGCTTTTTATATAACGACCGCAACGGGATTGTTTACAGGTTTCAGAAGGACGAAAAGACAGTACCTACTGATGCCGCTTGTCCCGTTGACATTCATCACTTTGTACTACTGGGACCTTGCATATGGGAATAAAGTGCATAGGATTCGAATGGAGGCGGAACACGTAATGACGCACGAAGCTGAATCCTTGGAGTGGCCGTGCGGTCTGCCTTCTCCCTCCTCCATCGACCTCGGACGATTCGACGAGGAGGAAAAGAAGAGAATACATCCGCCCCTTCTTCTCTAA
Protein
MGNLFSVSMEENYKRNQEFLQTINKVKMERQIQLQYQMQERQMALEIAKNRDTCLWITAFYITTATGLFTGFRRTKRQYLLMPLVPLTFITLYYWDLAYGNKVHRIRMEAEHVMTHEAESLEWPCGLPSPSSIDLGRFDEEEKKRIHPPLLL
Summary
Uniprot
A0A1E1WFI9
A0A2A4IZV3
A0A2H1VA21
A0A2W1BD19
A0A194QF76
A0A194QQW4
+ More
A0A212F7Q1 S4P716 A0A0L7L0Q3 A0A3S2M4K2 A0A067QN26 D6W8B5 A0A336M880 A0A1J1IBJ9 A0A1W4WKC8 A0A182G841 Q17KK6 A0A182J7S1 A0A182MZE8 A0A182QZM3 A0A182JPU6 A0A1S4HCB6 A0A182L2E5 A0A182ULZ4 A0A182X1L5 A0A182IEJ7 A0A182RVL2 A0A182SJ37 A0A182W7U2 A0A182F3W4 A0A182P0G2 A0A182TY69 A0A182M178 E0VME7 A0A2J7PXU6 B0W1N6 A0A3R7MZF5 A0A0L0BX10 A0A0P4W3N0 A0A0P4W047 A0A0P4VV09 E9HK56 A0A0P6HEC3 A0A1I8MN37 G8Z4Z0 V4BDI0 A0A0A1XBD4 A0A1I8Q6Y2 A0A2C9LPY0 A0A0K8VEE7 A0A034WYA2 T1GWL2 W8CA04 A0A1A9WVR1 A0A3S1C070 A0A2L2YUK2 A0A1A9VNH1 Q29H81 B4MTG2 B4L2T8 A0A0M4EU94 B3NVU3 B3MVS8 A0A1W4VTM3 Q9VY41 A0A2P2IA37 A0A3M6TJI2 Q4V627 A0A2R5LG94 B4JJ15 H3CH81 B4R4N9 Q4T1R7 A0A023FWP1 B4IGC4 B4M2W4 B4Q2S4 A0A3Q1B0C3 A0A3P8SGM8 A7RXB5 B3S3Z4 A0A2B4RB49 A0A3Q1FVP1 A0A3Q3XGX0 A0A0B6YI80 A0A1B0FQ65 A0A1B0AFC1 A0A2D0SYW1 A0A1B0AYL7 A0A3B4ZEV6 A0A3Q2YLK6 A0A1W4ZTT2 B9EP46 A0A060XLR7 A0A131Y1R1 A0A3P9CQ72 T1J1M9 E3TFX8 A0A2U9CEX1 A0A3B4BIQ1
A0A212F7Q1 S4P716 A0A0L7L0Q3 A0A3S2M4K2 A0A067QN26 D6W8B5 A0A336M880 A0A1J1IBJ9 A0A1W4WKC8 A0A182G841 Q17KK6 A0A182J7S1 A0A182MZE8 A0A182QZM3 A0A182JPU6 A0A1S4HCB6 A0A182L2E5 A0A182ULZ4 A0A182X1L5 A0A182IEJ7 A0A182RVL2 A0A182SJ37 A0A182W7U2 A0A182F3W4 A0A182P0G2 A0A182TY69 A0A182M178 E0VME7 A0A2J7PXU6 B0W1N6 A0A3R7MZF5 A0A0L0BX10 A0A0P4W3N0 A0A0P4W047 A0A0P4VV09 E9HK56 A0A0P6HEC3 A0A1I8MN37 G8Z4Z0 V4BDI0 A0A0A1XBD4 A0A1I8Q6Y2 A0A2C9LPY0 A0A0K8VEE7 A0A034WYA2 T1GWL2 W8CA04 A0A1A9WVR1 A0A3S1C070 A0A2L2YUK2 A0A1A9VNH1 Q29H81 B4MTG2 B4L2T8 A0A0M4EU94 B3NVU3 B3MVS8 A0A1W4VTM3 Q9VY41 A0A2P2IA37 A0A3M6TJI2 Q4V627 A0A2R5LG94 B4JJ15 H3CH81 B4R4N9 Q4T1R7 A0A023FWP1 B4IGC4 B4M2W4 B4Q2S4 A0A3Q1B0C3 A0A3P8SGM8 A7RXB5 B3S3Z4 A0A2B4RB49 A0A3Q1FVP1 A0A3Q3XGX0 A0A0B6YI80 A0A1B0FQ65 A0A1B0AFC1 A0A2D0SYW1 A0A1B0AYL7 A0A3B4ZEV6 A0A3Q2YLK6 A0A1W4ZTT2 B9EP46 A0A060XLR7 A0A131Y1R1 A0A3P9CQ72 T1J1M9 E3TFX8 A0A2U9CEX1 A0A3B4BIQ1
Pubmed
28756777
26354079
22118469
23622113
26227816
24845553
+ More
18362917 19820115 26483478 17510324 12364791 20966253 20566863 26108605 21292972 25315136 23254933 25830018 15562597 25348373 24495485 26561354 15632085 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 30382153 15496914 17550304 17615350 18719581 20433749 24755649 25186727 20634964 25463417
18362917 19820115 26483478 17510324 12364791 20966253 20566863 26108605 21292972 25315136 23254933 25830018 15562597 25348373 24495485 26561354 15632085 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 30382153 15496914 17550304 17615350 18719581 20433749 24755649 25186727 20634964 25463417
EMBL
GDQN01005336
JAT85718.1
NWSH01004790
PCG64682.1
ODYU01001261
SOQ37232.1
+ More
KZ150433 PZC70886.1 KQ459053 KPJ04198.1 KQ461181 KPJ07739.1 AGBW02009842 OWR49766.1 GAIX01006611 JAA85949.1 JTDY01003732 KOB69078.1 RSAL01000042 RVE50769.1 KK853224 KDR09687.1 KQ971307 EFA10888.2 UFQT01000461 SSX24577.1 CVRI01000043 CRK96358.1 JXUM01047381 KQ561516 KXJ78318.1 CH477223 EAT47220.1 AXCN02000606 AAAB01008964 APCN01005204 AXCM01006351 DS235307 EEB14553.1 NEVH01020852 PNF21168.1 DS231823 EDS26586.1 QCYY01002037 ROT73320.1 JRES01001303 KNC23774.1 GDRN01094818 JAI59660.1 GDRN01094822 GDRN01094821 JAI59658.1 GDRN01094819 JAI59659.1 GL732667 EFX67824.1 GDIQ01020440 JAN74297.1 GU186848 ADQ43240.1 KB203019 ESO86634.1 GBXI01006112 JAD08180.1 GDHF01015091 JAI37223.1 GAKP01000199 JAC58753.1 CAQQ02176559 GAMC01006201 JAC00355.1 RQTK01000454 RUS79388.1 IAAA01051843 LAA11783.1 CH379064 EAL31877.2 CH963851 EDW75401.2 CH933810 EDW06905.1 CP012528 ALC49288.1 CH954180 EDV47108.1 CH902625 EDV35073.1 AE014298 AAF48364.2 IACF01005194 LAB70780.1 RCHS01003481 RMX41501.1 BT022479 AAY54895.1 GGLE01004363 MBY08489.1 CH916370 EDW00556.1 CM000366 EDX17893.1 CAAE01010492 CAF93165.1 GBBL01002220 JAC25100.1 CH480835 EDW48858.1 CH940651 EDW65139.1 CM000162 EDX01668.2 DS469549 EDO44001.1 DS985249 EDV22364.1 LSMT01000793 PFX14356.1 HACG01008626 CEK55491.1 CCAG010006707 JXJN01005871 BT057421 ACM09293.1 FR905559 CDQ80172.1 GEFM01003965 JAP71831.1 JH431789 GU589263 ADO29214.1 CP026258 AWP15139.1 AWP15140.1 AWP15141.1 AWP15142.1 AWP15143.1
KZ150433 PZC70886.1 KQ459053 KPJ04198.1 KQ461181 KPJ07739.1 AGBW02009842 OWR49766.1 GAIX01006611 JAA85949.1 JTDY01003732 KOB69078.1 RSAL01000042 RVE50769.1 KK853224 KDR09687.1 KQ971307 EFA10888.2 UFQT01000461 SSX24577.1 CVRI01000043 CRK96358.1 JXUM01047381 KQ561516 KXJ78318.1 CH477223 EAT47220.1 AXCN02000606 AAAB01008964 APCN01005204 AXCM01006351 DS235307 EEB14553.1 NEVH01020852 PNF21168.1 DS231823 EDS26586.1 QCYY01002037 ROT73320.1 JRES01001303 KNC23774.1 GDRN01094818 JAI59660.1 GDRN01094822 GDRN01094821 JAI59658.1 GDRN01094819 JAI59659.1 GL732667 EFX67824.1 GDIQ01020440 JAN74297.1 GU186848 ADQ43240.1 KB203019 ESO86634.1 GBXI01006112 JAD08180.1 GDHF01015091 JAI37223.1 GAKP01000199 JAC58753.1 CAQQ02176559 GAMC01006201 JAC00355.1 RQTK01000454 RUS79388.1 IAAA01051843 LAA11783.1 CH379064 EAL31877.2 CH963851 EDW75401.2 CH933810 EDW06905.1 CP012528 ALC49288.1 CH954180 EDV47108.1 CH902625 EDV35073.1 AE014298 AAF48364.2 IACF01005194 LAB70780.1 RCHS01003481 RMX41501.1 BT022479 AAY54895.1 GGLE01004363 MBY08489.1 CH916370 EDW00556.1 CM000366 EDX17893.1 CAAE01010492 CAF93165.1 GBBL01002220 JAC25100.1 CH480835 EDW48858.1 CH940651 EDW65139.1 CM000162 EDX01668.2 DS469549 EDO44001.1 DS985249 EDV22364.1 LSMT01000793 PFX14356.1 HACG01008626 CEK55491.1 CCAG010006707 JXJN01005871 BT057421 ACM09293.1 FR905559 CDQ80172.1 GEFM01003965 JAP71831.1 JH431789 GU589263 ADO29214.1 CP026258 AWP15139.1 AWP15140.1 AWP15141.1 AWP15142.1 AWP15143.1
Proteomes
UP000218220
UP000053268
UP000053240
UP000007151
UP000037510
UP000283053
+ More
UP000027135 UP000007266 UP000183832 UP000192223 UP000069940 UP000249989 UP000008820 UP000075880 UP000075884 UP000075886 UP000075881 UP000075882 UP000075903 UP000076407 UP000075840 UP000075900 UP000075901 UP000075920 UP000069272 UP000075885 UP000075902 UP000075883 UP000009046 UP000235965 UP000002320 UP000283509 UP000037069 UP000000305 UP000095301 UP000030746 UP000095300 UP000076420 UP000015102 UP000091820 UP000271974 UP000078200 UP000001819 UP000007798 UP000009192 UP000092553 UP000008711 UP000007801 UP000192221 UP000000803 UP000275408 UP000001070 UP000007303 UP000000304 UP000001292 UP000008792 UP000002282 UP000257160 UP000265080 UP000001593 UP000009022 UP000225706 UP000257200 UP000261620 UP000092444 UP000092445 UP000221080 UP000092460 UP000261400 UP000264820 UP000192224 UP000087266 UP000193380 UP000265160 UP000246464 UP000261520
UP000027135 UP000007266 UP000183832 UP000192223 UP000069940 UP000249989 UP000008820 UP000075880 UP000075884 UP000075886 UP000075881 UP000075882 UP000075903 UP000076407 UP000075840 UP000075900 UP000075901 UP000075920 UP000069272 UP000075885 UP000075902 UP000075883 UP000009046 UP000235965 UP000002320 UP000283509 UP000037069 UP000000305 UP000095301 UP000030746 UP000095300 UP000076420 UP000015102 UP000091820 UP000271974 UP000078200 UP000001819 UP000007798 UP000009192 UP000092553 UP000008711 UP000007801 UP000192221 UP000000803 UP000275408 UP000001070 UP000007303 UP000000304 UP000001292 UP000008792 UP000002282 UP000257160 UP000265080 UP000001593 UP000009022 UP000225706 UP000257200 UP000261620 UP000092444 UP000092445 UP000221080 UP000092460 UP000261400 UP000264820 UP000192224 UP000087266 UP000193380 UP000265160 UP000246464 UP000261520
Pfam
PF10166 DUF2368
Interpro
IPR019319
Plg-R(KT)
ProteinModelPortal
A0A1E1WFI9
A0A2A4IZV3
A0A2H1VA21
A0A2W1BD19
A0A194QF76
A0A194QQW4
+ More
A0A212F7Q1 S4P716 A0A0L7L0Q3 A0A3S2M4K2 A0A067QN26 D6W8B5 A0A336M880 A0A1J1IBJ9 A0A1W4WKC8 A0A182G841 Q17KK6 A0A182J7S1 A0A182MZE8 A0A182QZM3 A0A182JPU6 A0A1S4HCB6 A0A182L2E5 A0A182ULZ4 A0A182X1L5 A0A182IEJ7 A0A182RVL2 A0A182SJ37 A0A182W7U2 A0A182F3W4 A0A182P0G2 A0A182TY69 A0A182M178 E0VME7 A0A2J7PXU6 B0W1N6 A0A3R7MZF5 A0A0L0BX10 A0A0P4W3N0 A0A0P4W047 A0A0P4VV09 E9HK56 A0A0P6HEC3 A0A1I8MN37 G8Z4Z0 V4BDI0 A0A0A1XBD4 A0A1I8Q6Y2 A0A2C9LPY0 A0A0K8VEE7 A0A034WYA2 T1GWL2 W8CA04 A0A1A9WVR1 A0A3S1C070 A0A2L2YUK2 A0A1A9VNH1 Q29H81 B4MTG2 B4L2T8 A0A0M4EU94 B3NVU3 B3MVS8 A0A1W4VTM3 Q9VY41 A0A2P2IA37 A0A3M6TJI2 Q4V627 A0A2R5LG94 B4JJ15 H3CH81 B4R4N9 Q4T1R7 A0A023FWP1 B4IGC4 B4M2W4 B4Q2S4 A0A3Q1B0C3 A0A3P8SGM8 A7RXB5 B3S3Z4 A0A2B4RB49 A0A3Q1FVP1 A0A3Q3XGX0 A0A0B6YI80 A0A1B0FQ65 A0A1B0AFC1 A0A2D0SYW1 A0A1B0AYL7 A0A3B4ZEV6 A0A3Q2YLK6 A0A1W4ZTT2 B9EP46 A0A060XLR7 A0A131Y1R1 A0A3P9CQ72 T1J1M9 E3TFX8 A0A2U9CEX1 A0A3B4BIQ1
A0A212F7Q1 S4P716 A0A0L7L0Q3 A0A3S2M4K2 A0A067QN26 D6W8B5 A0A336M880 A0A1J1IBJ9 A0A1W4WKC8 A0A182G841 Q17KK6 A0A182J7S1 A0A182MZE8 A0A182QZM3 A0A182JPU6 A0A1S4HCB6 A0A182L2E5 A0A182ULZ4 A0A182X1L5 A0A182IEJ7 A0A182RVL2 A0A182SJ37 A0A182W7U2 A0A182F3W4 A0A182P0G2 A0A182TY69 A0A182M178 E0VME7 A0A2J7PXU6 B0W1N6 A0A3R7MZF5 A0A0L0BX10 A0A0P4W3N0 A0A0P4W047 A0A0P4VV09 E9HK56 A0A0P6HEC3 A0A1I8MN37 G8Z4Z0 V4BDI0 A0A0A1XBD4 A0A1I8Q6Y2 A0A2C9LPY0 A0A0K8VEE7 A0A034WYA2 T1GWL2 W8CA04 A0A1A9WVR1 A0A3S1C070 A0A2L2YUK2 A0A1A9VNH1 Q29H81 B4MTG2 B4L2T8 A0A0M4EU94 B3NVU3 B3MVS8 A0A1W4VTM3 Q9VY41 A0A2P2IA37 A0A3M6TJI2 Q4V627 A0A2R5LG94 B4JJ15 H3CH81 B4R4N9 Q4T1R7 A0A023FWP1 B4IGC4 B4M2W4 B4Q2S4 A0A3Q1B0C3 A0A3P8SGM8 A7RXB5 B3S3Z4 A0A2B4RB49 A0A3Q1FVP1 A0A3Q3XGX0 A0A0B6YI80 A0A1B0FQ65 A0A1B0AFC1 A0A2D0SYW1 A0A1B0AYL7 A0A3B4ZEV6 A0A3Q2YLK6 A0A1W4ZTT2 B9EP46 A0A060XLR7 A0A131Y1R1 A0A3P9CQ72 T1J1M9 E3TFX8 A0A2U9CEX1 A0A3B4BIQ1
Ontologies
PANTHER
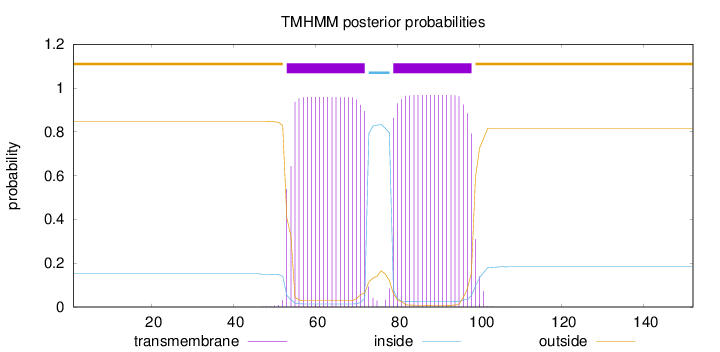
Topology
Length:
152
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
38.03611
Exp number, first 60 AAs:
6.95889
Total prob of N-in:
0.15207
outside
1 - 52
TMhelix
53 - 72
inside
73 - 78
TMhelix
79 - 98
outside
99 - 152
Population Genetic Test Statistics
Pi
308.519758
Theta
175.504746
Tajima's D
2.208435
CLR
0.100384
CSRT
0.911404429778511
Interpretation
Uncertain
