Gene
KWMTBOMO12174
Pre Gene Modal
BGIBMGA004222
Annotation
PREDICTED:_extracellular_signal-regulated_kinase_2_[Bombyx_mori]
Full name
Mitogen-activated protein kinase
Location in the cell
Cytoplasmic Reliability : 1.744 Nuclear Reliability : 1.684
Sequence
CDS
ATGAATCAGAACCAGAAAAGAGATGACAAGAAGAAGGACGGACATAAGCATCAACAGAAAAAGAGCTCGGAAAAAAATATGTCCGAAATAGACGAGCACATATTAAAGAGATTCGAAATTAAAAAGAGATTGGGCAAAGGGGCCTACGGGATTGTTTGGAAGGCTGTCGACAAGAAAACCAAAGACGTGGTCGCAATAAAAAAGATATTTGACGCGTTCAGAAACCAGACCGACGCTCAGAGGACGTTCCGGGAGATTATATTCTTGCAATCGTTCAGAAATCACCCGAACATTGTTAAACTACACAGCATACATAGAGCTCTGAACAACAACGACATATATCTGGGATTCGAATACATGGAAACGGACCTGCATAACGTCATCAAACGCGGCAACATACTGAAAGACATCCACAAACGGTACATAATGTATCAAATGCTTAAAGCGACGAAGTACATACACTCTGGGAACGTGATACATCGCGATCAAAAGCCCAGTAACGTGTTAATAGACAGCGCTTGTAGAGTGAAGTTGGCAGATTTCGGTCTGGCTCGCTCAGTCTCTAGTGTGTACTCTGGCGGAGAAGATGGCGCAGATCCTTGTCTGACTGATTACGTCGCCACGCGCTGGTACCGAGCCCCGGAGATACTTATAGCTAGCAAGAATTACACAAAAGGAATCGACATGTGGTCCCTTGGTTGTATCCTGGCTGAGATGCTGGTTGGGGAGCCTCTGTTTCCCGGGACATCGACCGTCAATCAAATTGAGAGGATCATGGCGACACTCCCGAGGCCATCGAACGAAGGTGAGCAACAGAATTTTAAATTTTTTATTGCTTAA
Protein
MNQNQKRDDKKKDGHKHQQKKSSEKNMSEIDEHILKRFEIKKRLGKGAYGIVWKAVDKKTKDVVAIKKIFDAFRNQTDAQRTFREIIFLQSFRNHPNIVKLHSIHRALNNNDIYLGFEYMETDLHNVIKRGNILKDIHKRYIMYQMLKATKYIHSGNVIHRDQKPSNVLIDSACRVKLADFGLARSVSSVYSGGEDGADPCLTDYVATRWYRAPEILIASKNYTKGIDMWSLGCILAEMLVGEPLFPGTSTVNQIERIMATLPRPSNEGEQQNFKFFIA
Summary
Catalytic Activity
ATP + L-seryl-[protein] = ADP + H(+) + O-phospho-L-seryl-[protein]
ATP + L-threonyl-[protein] = ADP + H(+) + O-phospho-L-threonyl-[protein]
ATP + L-threonyl-[protein] = ADP + H(+) + O-phospho-L-threonyl-[protein]
Cofactor
Mg(2+)
Similarity
Belongs to the protein kinase superfamily. Ser/Thr protein kinase family. MAP kinase subfamily.
Feature
chain Mitogen-activated protein kinase
Uniprot
H9J3Y3
A0A3S2TNB7
A0A194QQF2
A0A194QFD7
A0A2A4J1G8
A0A212F7P9
+ More
A0A1Y1KUB7 A0A0T6BE67 A0A2W1BC41 A0A1W4WSE0 A0A195B782 A0A067RR94 E0VH91 D6WRF3 A0A1B6JV20 E9FW56 A0A267GKN5 K7J0J0 A0A1B6K2A3 A0A0N8DX77 C3YL59 A0A1I8IET1 A0A3L8D757 A0A1I8FX20 V3ZBB1 A0A1I8G9P2 A0A0P5B6P5 A0A0P5B5D1 A0A0C9PSH0 A0A1B6L5P6 A0A0P4ZJK7 A0A1B6FR54 A0A151IMN8 A0A0P4XMK0 A0A151IS01 A0A0P5BC95 A0A0P5BG15 A0A0P5EXJ4 E2A9I8 A0A158NHA2 A0A0P5FUV4 A0A0P5G8C0 A0A267DXK0 A0A0P5QP46 A0A0P5TUJ7 A0A267EB49 A0A0P6I8K5 A0A2J7QYA8 A0A0P5TG99 A0A2J7QY94 C3ZCU0 A0A0R4IIR5 Q503Q2 A0A1S3JU08 A0A0P6AYG3 R7UWR1 A0A2J7QYA0 A0A0K8UX06 A0A1B6DKB0 A0A210PNM3 A0A3Q0RUQ4 A0A1I8P293 A0A3B5AQG8 A0A1I8P2E0 A0A1I8IFA0 A0A2C9JR00 A0A3B3QHF8 G3NNS3 A0A3B3QGG0 A0A0M8ZZY4 A0A3M6UF61 A0A2D0PYT4 A0A1W4ZQQ6 A0A154PS86 A0A2B4SJ31 I3J2P3 A0A0P7ZGZ3 A0A1S3Q1T8 A0A3Q2WL22 A0A3Q3E9N2 A0A3P9B7J6 A0A3P8Q727 A0A0L0BXI8 A0A1W4ZZV0 A0A3Q3S8C4 A0A3B4XX16 A0A3P8WN64 A0A3B4D0J0 A0A087ZZ64 W5MPZ7 A0A3B1JNL5 A0A2A3EH65 A0A3P9KA90 A0A345DKR8 A0A1S3Q1U6 A0A3Q1IFL4 A0A1I8JIJ6 A0A0L8FSR6 A0A1I8I6G0 A0A288W1R9
A0A1Y1KUB7 A0A0T6BE67 A0A2W1BC41 A0A1W4WSE0 A0A195B782 A0A067RR94 E0VH91 D6WRF3 A0A1B6JV20 E9FW56 A0A267GKN5 K7J0J0 A0A1B6K2A3 A0A0N8DX77 C3YL59 A0A1I8IET1 A0A3L8D757 A0A1I8FX20 V3ZBB1 A0A1I8G9P2 A0A0P5B6P5 A0A0P5B5D1 A0A0C9PSH0 A0A1B6L5P6 A0A0P4ZJK7 A0A1B6FR54 A0A151IMN8 A0A0P4XMK0 A0A151IS01 A0A0P5BC95 A0A0P5BG15 A0A0P5EXJ4 E2A9I8 A0A158NHA2 A0A0P5FUV4 A0A0P5G8C0 A0A267DXK0 A0A0P5QP46 A0A0P5TUJ7 A0A267EB49 A0A0P6I8K5 A0A2J7QYA8 A0A0P5TG99 A0A2J7QY94 C3ZCU0 A0A0R4IIR5 Q503Q2 A0A1S3JU08 A0A0P6AYG3 R7UWR1 A0A2J7QYA0 A0A0K8UX06 A0A1B6DKB0 A0A210PNM3 A0A3Q0RUQ4 A0A1I8P293 A0A3B5AQG8 A0A1I8P2E0 A0A1I8IFA0 A0A2C9JR00 A0A3B3QHF8 G3NNS3 A0A3B3QGG0 A0A0M8ZZY4 A0A3M6UF61 A0A2D0PYT4 A0A1W4ZQQ6 A0A154PS86 A0A2B4SJ31 I3J2P3 A0A0P7ZGZ3 A0A1S3Q1T8 A0A3Q2WL22 A0A3Q3E9N2 A0A3P9B7J6 A0A3P8Q727 A0A0L0BXI8 A0A1W4ZZV0 A0A3Q3S8C4 A0A3B4XX16 A0A3P8WN64 A0A3B4D0J0 A0A087ZZ64 W5MPZ7 A0A3B1JNL5 A0A2A3EH65 A0A3P9KA90 A0A345DKR8 A0A1S3Q1U6 A0A3Q1IFL4 A0A1I8JIJ6 A0A0L8FSR6 A0A1I8I6G0 A0A288W1R9
EC Number
2.7.11.24
Pubmed
EMBL
BABH01040234
RSAL01000042
RVE50768.1
KQ461181
KPJ07738.1
KQ459053
+ More
KPJ04197.1 NWSH01003771 PCG65831.1 AGBW02009842 OWR49765.1 GEZM01077409 JAV63315.1 LJIG01001733 KRT85207.1 KZ150433 PZC70887.1 KQ976574 KYM80115.1 KK852513 KDR22274.1 DS235165 EEB12747.1 KQ971351 EFA06559.2 GECU01004648 JAT03059.1 GL732525 EFX88665.1 NIVC01000272 PAA86593.1 GECU01002413 JAT05294.1 GDIQ01083052 JAN11685.1 GG666525 EEN59050.1 QOIP01000012 RLU16305.1 KB202719 ESO88318.1 GDIP01189375 JAJ34027.1 GDIP01189374 JAJ34028.1 GBYB01004273 JAG74040.1 GEBQ01020960 JAT19017.1 GDIP01213382 JAJ10020.1 GECZ01017077 JAS52692.1 KQ977035 KYN06191.1 GDIP01240780 LRGB01000024 JAI82621.1 KZS21579.1 KQ981098 KYN09495.1 GDIP01191821 JAJ31581.1 GDIP01189376 JAJ34026.1 GDIQ01266364 JAJ85360.1 GL437917 EFN69878.1 ADTU01015417 GDIQ01251238 JAK00487.1 GDIQ01248087 JAK03638.1 NIVC01003101 PAA53347.1 GDIQ01117735 JAL33991.1 GDIP01126629 JAL77085.1 NIVC01002421 PAA58029.1 GDIQ01008015 JAN86722.1 NEVH01009118 PNF33559.1 GDIP01128072 JAL75642.1 PNF33560.1 GG666610 EEN49593.1 CU633987 BC095227 BC165603 AAH95227.1 AAI65603.1 GDIP01022915 JAM80800.1 AMQN01005968 KB297235 ELU10754.1 PNF33558.1 GDHF01023633 GDHF01021193 GDHF01014970 JAI28681.1 JAI31121.1 JAI37344.1 GEDC01011165 JAS26133.1 NEDP02005573 OWF38077.1 KQ435804 KOX73063.1 RCHS01001704 RMX52058.1 KQ435037 KZC14128.1 LSMT01000082 PFX28482.1 AERX01011176 JARO02000009 KPP80295.1 JRES01001179 KNC24768.1 AHAT01020214 KZ288262 PBC30351.1 MF802853 AXF94777.1 KQ426816 KOF67724.1 KX424625 ARJ54258.1
KPJ04197.1 NWSH01003771 PCG65831.1 AGBW02009842 OWR49765.1 GEZM01077409 JAV63315.1 LJIG01001733 KRT85207.1 KZ150433 PZC70887.1 KQ976574 KYM80115.1 KK852513 KDR22274.1 DS235165 EEB12747.1 KQ971351 EFA06559.2 GECU01004648 JAT03059.1 GL732525 EFX88665.1 NIVC01000272 PAA86593.1 GECU01002413 JAT05294.1 GDIQ01083052 JAN11685.1 GG666525 EEN59050.1 QOIP01000012 RLU16305.1 KB202719 ESO88318.1 GDIP01189375 JAJ34027.1 GDIP01189374 JAJ34028.1 GBYB01004273 JAG74040.1 GEBQ01020960 JAT19017.1 GDIP01213382 JAJ10020.1 GECZ01017077 JAS52692.1 KQ977035 KYN06191.1 GDIP01240780 LRGB01000024 JAI82621.1 KZS21579.1 KQ981098 KYN09495.1 GDIP01191821 JAJ31581.1 GDIP01189376 JAJ34026.1 GDIQ01266364 JAJ85360.1 GL437917 EFN69878.1 ADTU01015417 GDIQ01251238 JAK00487.1 GDIQ01248087 JAK03638.1 NIVC01003101 PAA53347.1 GDIQ01117735 JAL33991.1 GDIP01126629 JAL77085.1 NIVC01002421 PAA58029.1 GDIQ01008015 JAN86722.1 NEVH01009118 PNF33559.1 GDIP01128072 JAL75642.1 PNF33560.1 GG666610 EEN49593.1 CU633987 BC095227 BC165603 AAH95227.1 AAI65603.1 GDIP01022915 JAM80800.1 AMQN01005968 KB297235 ELU10754.1 PNF33558.1 GDHF01023633 GDHF01021193 GDHF01014970 JAI28681.1 JAI31121.1 JAI37344.1 GEDC01011165 JAS26133.1 NEDP02005573 OWF38077.1 KQ435804 KOX73063.1 RCHS01001704 RMX52058.1 KQ435037 KZC14128.1 LSMT01000082 PFX28482.1 AERX01011176 JARO02000009 KPP80295.1 JRES01001179 KNC24768.1 AHAT01020214 KZ288262 PBC30351.1 MF802853 AXF94777.1 KQ426816 KOF67724.1 KX424625 ARJ54258.1
Proteomes
UP000005204
UP000283053
UP000053240
UP000053268
UP000218220
UP000007151
+ More
UP000192223 UP000078540 UP000027135 UP000009046 UP000007266 UP000000305 UP000215902 UP000002358 UP000001554 UP000095280 UP000279307 UP000030746 UP000078542 UP000076858 UP000078492 UP000000311 UP000005205 UP000235965 UP000000437 UP000085678 UP000014760 UP000242188 UP000261340 UP000095300 UP000261400 UP000076420 UP000261540 UP000007635 UP000053105 UP000275408 UP000221080 UP000192224 UP000076502 UP000225706 UP000005207 UP000034805 UP000087266 UP000264840 UP000261660 UP000265160 UP000265100 UP000037069 UP000261640 UP000261360 UP000265120 UP000261440 UP000005203 UP000018468 UP000018467 UP000242457 UP000265180 UP000265040 UP000053454
UP000192223 UP000078540 UP000027135 UP000009046 UP000007266 UP000000305 UP000215902 UP000002358 UP000001554 UP000095280 UP000279307 UP000030746 UP000078542 UP000076858 UP000078492 UP000000311 UP000005205 UP000235965 UP000000437 UP000085678 UP000014760 UP000242188 UP000261340 UP000095300 UP000261400 UP000076420 UP000261540 UP000007635 UP000053105 UP000275408 UP000221080 UP000192224 UP000076502 UP000225706 UP000005207 UP000034805 UP000087266 UP000264840 UP000261660 UP000265160 UP000265100 UP000037069 UP000261640 UP000261360 UP000265120 UP000261440 UP000005203 UP000018468 UP000018467 UP000242457 UP000265180 UP000265040 UP000053454
PRIDE
Interpro
SUPFAM
SSF56112
SSF56112
ProteinModelPortal
H9J3Y3
A0A3S2TNB7
A0A194QQF2
A0A194QFD7
A0A2A4J1G8
A0A212F7P9
+ More
A0A1Y1KUB7 A0A0T6BE67 A0A2W1BC41 A0A1W4WSE0 A0A195B782 A0A067RR94 E0VH91 D6WRF3 A0A1B6JV20 E9FW56 A0A267GKN5 K7J0J0 A0A1B6K2A3 A0A0N8DX77 C3YL59 A0A1I8IET1 A0A3L8D757 A0A1I8FX20 V3ZBB1 A0A1I8G9P2 A0A0P5B6P5 A0A0P5B5D1 A0A0C9PSH0 A0A1B6L5P6 A0A0P4ZJK7 A0A1B6FR54 A0A151IMN8 A0A0P4XMK0 A0A151IS01 A0A0P5BC95 A0A0P5BG15 A0A0P5EXJ4 E2A9I8 A0A158NHA2 A0A0P5FUV4 A0A0P5G8C0 A0A267DXK0 A0A0P5QP46 A0A0P5TUJ7 A0A267EB49 A0A0P6I8K5 A0A2J7QYA8 A0A0P5TG99 A0A2J7QY94 C3ZCU0 A0A0R4IIR5 Q503Q2 A0A1S3JU08 A0A0P6AYG3 R7UWR1 A0A2J7QYA0 A0A0K8UX06 A0A1B6DKB0 A0A210PNM3 A0A3Q0RUQ4 A0A1I8P293 A0A3B5AQG8 A0A1I8P2E0 A0A1I8IFA0 A0A2C9JR00 A0A3B3QHF8 G3NNS3 A0A3B3QGG0 A0A0M8ZZY4 A0A3M6UF61 A0A2D0PYT4 A0A1W4ZQQ6 A0A154PS86 A0A2B4SJ31 I3J2P3 A0A0P7ZGZ3 A0A1S3Q1T8 A0A3Q2WL22 A0A3Q3E9N2 A0A3P9B7J6 A0A3P8Q727 A0A0L0BXI8 A0A1W4ZZV0 A0A3Q3S8C4 A0A3B4XX16 A0A3P8WN64 A0A3B4D0J0 A0A087ZZ64 W5MPZ7 A0A3B1JNL5 A0A2A3EH65 A0A3P9KA90 A0A345DKR8 A0A1S3Q1U6 A0A3Q1IFL4 A0A1I8JIJ6 A0A0L8FSR6 A0A1I8I6G0 A0A288W1R9
A0A1Y1KUB7 A0A0T6BE67 A0A2W1BC41 A0A1W4WSE0 A0A195B782 A0A067RR94 E0VH91 D6WRF3 A0A1B6JV20 E9FW56 A0A267GKN5 K7J0J0 A0A1B6K2A3 A0A0N8DX77 C3YL59 A0A1I8IET1 A0A3L8D757 A0A1I8FX20 V3ZBB1 A0A1I8G9P2 A0A0P5B6P5 A0A0P5B5D1 A0A0C9PSH0 A0A1B6L5P6 A0A0P4ZJK7 A0A1B6FR54 A0A151IMN8 A0A0P4XMK0 A0A151IS01 A0A0P5BC95 A0A0P5BG15 A0A0P5EXJ4 E2A9I8 A0A158NHA2 A0A0P5FUV4 A0A0P5G8C0 A0A267DXK0 A0A0P5QP46 A0A0P5TUJ7 A0A267EB49 A0A0P6I8K5 A0A2J7QYA8 A0A0P5TG99 A0A2J7QY94 C3ZCU0 A0A0R4IIR5 Q503Q2 A0A1S3JU08 A0A0P6AYG3 R7UWR1 A0A2J7QYA0 A0A0K8UX06 A0A1B6DKB0 A0A210PNM3 A0A3Q0RUQ4 A0A1I8P293 A0A3B5AQG8 A0A1I8P2E0 A0A1I8IFA0 A0A2C9JR00 A0A3B3QHF8 G3NNS3 A0A3B3QGG0 A0A0M8ZZY4 A0A3M6UF61 A0A2D0PYT4 A0A1W4ZQQ6 A0A154PS86 A0A2B4SJ31 I3J2P3 A0A0P7ZGZ3 A0A1S3Q1T8 A0A3Q2WL22 A0A3Q3E9N2 A0A3P9B7J6 A0A3P8Q727 A0A0L0BXI8 A0A1W4ZZV0 A0A3Q3S8C4 A0A3B4XX16 A0A3P8WN64 A0A3B4D0J0 A0A087ZZ64 W5MPZ7 A0A3B1JNL5 A0A2A3EH65 A0A3P9KA90 A0A345DKR8 A0A1S3Q1U6 A0A3Q1IFL4 A0A1I8JIJ6 A0A0L8FSR6 A0A1I8I6G0 A0A288W1R9
PDB
3OZ6
E-value=6.03975e-77,
Score=730
Ontologies
GO
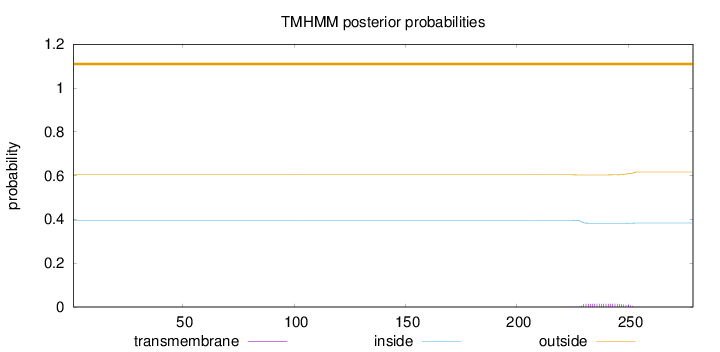
Topology
Length:
279
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.31387
Exp number, first 60 AAs:
0
Total prob of N-in:
0.39648
outside
1 - 279
Population Genetic Test Statistics
Pi
302.651714
Theta
160.254354
Tajima's D
2.010877
CLR
0.271487
CSRT
0.88155592220389
Interpretation
Uncertain
