Gene
KWMTBOMO12166 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA014059
Annotation
PREDICTED:_NADH_dehydrogenase_[ubiquinone]_1_alpha_subcomplex_subunit_10?_mitochondrial_[Papilio_polytes]
Full name
NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 10, mitochondrial
Alternative Name
Complex I-42kD
NADH-ubiquinone oxidoreductase 42 kDa subunit
NADH-ubiquinone oxidoreductase 42 kDa subunit
Location in the cell
Cytoplasmic Reliability : 1.597 Mitochondrial Reliability : 1.615
Sequence
CDS
ATGGCTACTCTCGTCAGAACAGCTGTAATTAAAGTTATCAGCCCAACTCAGGGTACAAAAATAGCAGCATGTACGTTTGTACAAAATAGGAATATCTCCGGGCGGGCCATGCGGGAAGCGCTACCACCTCCTCCACCGAAACCTGCACCCTTTGACTATGTGAACAAGGACTACACTTGGTTGCGCAGCTTAATCGACCGCACGACCCACAGGTTTGATGATAACAGCAAAGTGATAATCGTGGAAGGGCCGGTAGCTGCTGGTAAGACCGCCTTTGCAGCCTCCCTAGCCGATGACTTGGGTATGAAGCATTTCCCCGAAGCTAACATGGATTTGCACTATATTCGTCCGAACGGCGTCGACTTGAGGTCCTTCGACGACCAAGTACCCGAGGACACTAGGACCTTTGACCATGTTAATTTCAATCAGAATCCTAACCATCGCTTGGCTGCAAACTTCCAAATAATGATGTATGTGGCCCGGTACAGTCAGTATATTGATGCTTTGGCACACCTGTTCAATACTGGCCAAGGAATTGTGTTGGAGAGGTCCCCTTACTCTGACTTTGTATTCTTGGAAGCTATGTATTCTCAGAAATTCTTGAGTAAGGCAGTCAGGTCAACTTACTATGAGCTGCGAGAAAACACAATTGAGGAGCTAATGCGTCCACATCTAGTTATTTATCTGGATCTTCCTGTTTCCAAGGTTCAAGAAGCTATTAAAAAACGTGCTCTCAGTCACGAGGTGACCAGCAAAGCTCTCACACCGGCCTTCCTGACTGAAATTGAGAAGCAGTACAAAAACAAGTATCTGAGAGATATCGCCACTCATGCAGAGCTGCTCGTGTACGACTGGACCGGAGGAGGTGAAGTCGAAGTGGTCGTCGAGGACATAGAGCGACTGAACTTCGACCAGTATACGGAGCGTGAAGAGCCCAAGATGAAGGACTGGCGTTTGCCTCGCGAAGTGGACTGGGCCGACCAGCGCGCGCTCTACACGAACCAGAAGCACTTGCTGATGAATCTGCTCGCCATCCCCAAACTGGACATACCGGAGCTGATAACTAGCGCCGAGGACTGTTACGAGAGAGAAAAGGTTATTTCTGCTCACCCAGCCTTCCGTTACGAACTGGGATACGCGCCAGGCGAATCTGTGACCTTCAAGAACAAGCTGCCCAAATACCACGAGTACATATAG
Protein
MATLVRTAVIKVISPTQGTKIAACTFVQNRNISGRAMREALPPPPPKPAPFDYVNKDYTWLRSLIDRTTHRFDDNSKVIIVEGPVAAGKTAFAASLADDLGMKHFPEANMDLHYIRPNGVDLRSFDDQVPEDTRTFDHVNFNQNPNHRLAANFQIMMYVARYSQYIDALAHLFNTGQGIVLERSPYSDFVFLEAMYSQKFLSKAVRSTYYELRENTIEELMRPHLVIYLDLPVSKVQEAIKKRALSHEVTSKALTPAFLTEIEKQYKNKYLRDIATHAELLVYDWTGGGEVEVVVEDIERLNFDQYTEREEPKMKDWRLPREVDWADQRALYTNQKHLLMNLLAIPKLDIPELITSAEDCYEREKVISAHPAFRYELGYAPGESVTFKNKLPKYHEYI
Summary
Description
Accessory subunit of the mitochondrial membrane respiratory chain NADH dehydrogenase (Complex I), that is believed not to be involved in catalysis. Complex I functions in the transfer of electrons from NADH to the respiratory chain. The immediate electron acceptor for the enzyme is believed to be ubiquinone.
Accessory subunit of the mitochondrial membrane respiratory chain NADH dehydrogenase (Complex I), that is believed not to be involved in catalysis. Complex I functions in the transfer of electrons from NADH to the respiratory chain. The immediate electron acceptor for the enzyme is believed to be ubiquinone (By similarity).
Accessory subunit of the mitochondrial membrane respiratory chain NADH dehydrogenase (Complex I), that is believed not to be involved in catalysis. Complex I functions in the transfer of electrons from NADH to the respiratory chain. The immediate electron acceptor for the enzyme is believed to be ubiquinone (By similarity).
Cofactor
FAD
Subunit
Complex I is composed of 45 different subunits. This a component of the hydrophobic protein fraction (By similarity).
Similarity
Belongs to the complex I NDUFA10 subunit family.
Keywords
Complete proteome
Electron transport
FAD
Flavoprotein
Mitochondrion
Reference proteome
Respiratory chain
Transit peptide
Transport
Feature
chain NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 10, mitochondrial
Uniprot
H9JWZ4
A0A2W1BFM8
S4PPT2
I4DJN1
A0A2A4J4B3
A0A2H1VMP4
+ More
A0A3S2LPC2 A0A212F375 A0A220K8N4 A0A0L7L2W1 A0A0N1IKH2 A0A0K8TSV4 B4NHR3 A0A1B6I241 B3LVL1 A0A1W4VQ24 B4M0N2 A0A0L0C5X1 B4K6L1 A0A1L8EDU1 A0A1L8EDU6 A0A1J1ICX8 A0A0M4F600 B4QZD1 B4HM81 B4JRL6 B3P7P1 A0A034W901 Q297T7 B0WW92 A4V383 P91929 A0A0K8VIR8 A0A1B6LQC9 B4PLJ6 A0A1I8Q2W2 A0A182M9V2 A0A3B0JFE9 B4G309 A0A182WDL5 A0A182YAZ9 D3TLY4 A0A067R4C9 A0A2M3Z1B4 Q7QHC7 A0A182TDY3 A0A2J7QKF4 A0A2M4BRB2 T1PBJ6 A0A2M3ZG26 W8BR54 A0A1Q3FH78 A0A1I8MID9 A0A182X5Q2 W5JP72 A0A1A9YQF0 A0A182VAU3 T1E8C6 A0A182HW95 A0A1B0AY20 A0A182FW35 A0A182KL06 D1ZZE8 A0A182NZQ0 A0A182Q7B1 A0A2M4A7D5 A0A182T4L6 A0A182N286 A0A1B0A1A9 A0A182RW40 A0A023F1W2 A0A084WM05 A0A182JX01 A0A224XRJ6 A0A1L8E0J4 A0A1A9VWM7 U5EVK8 A0A023ERK6 V5G650 A0A182G813 Q1HQV6 A0A182JDF2 A0A1B6FVR3 A0A1B0D7X3 A0A1A9WWW0 A0A1B6DH01 A0A3R7SKG8 A0A0T6B1G8 T1HTY7 A0A1B0CQN1 A0A1B0D3Z1 J3JVZ9 U4UML3 N6U8T6 J9K3B7 A0A170ZEA3 A0A2H8U1Y7 A0A146ME71 A0A1W4XC65
A0A3S2LPC2 A0A212F375 A0A220K8N4 A0A0L7L2W1 A0A0N1IKH2 A0A0K8TSV4 B4NHR3 A0A1B6I241 B3LVL1 A0A1W4VQ24 B4M0N2 A0A0L0C5X1 B4K6L1 A0A1L8EDU1 A0A1L8EDU6 A0A1J1ICX8 A0A0M4F600 B4QZD1 B4HM81 B4JRL6 B3P7P1 A0A034W901 Q297T7 B0WW92 A4V383 P91929 A0A0K8VIR8 A0A1B6LQC9 B4PLJ6 A0A1I8Q2W2 A0A182M9V2 A0A3B0JFE9 B4G309 A0A182WDL5 A0A182YAZ9 D3TLY4 A0A067R4C9 A0A2M3Z1B4 Q7QHC7 A0A182TDY3 A0A2J7QKF4 A0A2M4BRB2 T1PBJ6 A0A2M3ZG26 W8BR54 A0A1Q3FH78 A0A1I8MID9 A0A182X5Q2 W5JP72 A0A1A9YQF0 A0A182VAU3 T1E8C6 A0A182HW95 A0A1B0AY20 A0A182FW35 A0A182KL06 D1ZZE8 A0A182NZQ0 A0A182Q7B1 A0A2M4A7D5 A0A182T4L6 A0A182N286 A0A1B0A1A9 A0A182RW40 A0A023F1W2 A0A084WM05 A0A182JX01 A0A224XRJ6 A0A1L8E0J4 A0A1A9VWM7 U5EVK8 A0A023ERK6 V5G650 A0A182G813 Q1HQV6 A0A182JDF2 A0A1B6FVR3 A0A1B0D7X3 A0A1A9WWW0 A0A1B6DH01 A0A3R7SKG8 A0A0T6B1G8 T1HTY7 A0A1B0CQN1 A0A1B0D3Z1 J3JVZ9 U4UML3 N6U8T6 J9K3B7 A0A170ZEA3 A0A2H8U1Y7 A0A146ME71 A0A1W4XC65
Pubmed
19121390
28756777
23622113
22651552
26354079
22118469
+ More
28630352 26227816 26369729 17994087 26108605 25348373 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 12537569 10071211 17550304 25244985 20353571 24845553 12364791 14747013 17210077 24495485 25315136 20920257 23761445 20966253 18362917 19820115 25474469 24438588 24945155 26483478 17204158 17510324 22516182 23537049 26823975
28630352 26227816 26369729 17994087 26108605 25348373 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 12537569 10071211 17550304 25244985 20353571 24845553 12364791 14747013 17210077 24495485 25315136 20920257 23761445 20966253 18362917 19820115 25474469 24438588 24945155 26483478 17204158 17510324 22516182 23537049 26823975
EMBL
BABH01041746
KZ150228
PZC72027.1
GAIX01000027
JAA92533.1
AK401499
+ More
KQ459053 BAM18121.1 KPJ04189.1 NWSH01003461 PCG66253.1 ODYU01003385 SOQ42077.1 RSAL01000495 RVE41533.1 AGBW02010605 OWR48187.1 MF319745 ASJ26459.1 JTDY01003379 KOB69646.1 LADJ01000094 KPJ20830.1 GDAI01000149 JAI17454.1 CH964272 EDW84673.1 GECU01026713 JAS80993.1 CH902617 EDV42581.1 CH940650 EDW67324.1 JRES01000955 KNC26814.1 CH933806 EDW16311.1 GFDG01001973 JAV16826.1 GFDG01001974 JAV16825.1 CVRI01000047 CRK98059.1 CP012526 ALC47400.1 CM000364 EDX13867.1 CH480815 EDW43129.1 CH916373 EDV94406.1 CH954182 EDV54202.1 GAKP01008179 GAKP01008178 GAKP01008177 GAKP01008175 GAKP01008173 JAC50777.1 CM000070 EAL28118.1 DS232140 EDS35924.1 AE014297 AAN13899.1 AY051764 Y10911 GDHF01031881 GDHF01027384 GDHF01021211 GDHF01013561 GDHF01011070 GDHF01001202 JAI20433.1 JAI24930.1 JAI31103.1 JAI38753.1 JAI41244.1 JAI51112.1 GEBQ01014124 JAT25853.1 CM000160 EDW97945.1 AXCM01004806 OUUW01000005 SPP81057.1 CH479179 EDW24204.1 CCAG010001866 EZ422436 ADD18712.1 KK852902 KDR14073.1 GGFM01001520 MBW22271.1 AAAB01008816 EAA05289.4 NEVH01013262 PNF29067.1 GGFJ01006167 MBW55308.1 KA646141 AFP60770.1 GGFM01006731 MBW27482.1 GAMC01005003 JAC01553.1 GFDL01008137 JAV26908.1 ADMH02000909 ETN64695.1 GAMD01002888 JAA98702.1 APCN01001478 JXJN01005495 KQ971338 EFA02878.1 AXCN02001731 GGFK01003331 MBW36652.1 GBBI01003389 JAC15323.1 ATLV01024337 KE525351 KFB51249.1 GFTR01005827 JAW10599.1 GFDF01001863 JAV12221.1 GANO01001834 JAB58037.1 GAPW01001785 JAC11813.1 GALX01002921 JAB65545.1 JXUM01151271 KQ571179 KXJ68229.1 DQ440338 CH477951 ABF18371.1 EAT34841.1 GECZ01015530 JAS54239.1 AJVK01032324 GEDC01012329 JAS24969.1 QCYY01003201 ROT64718.1 LJIG01016233 KRT81217.1 ACPB03011673 AJWK01023776 AJVK01023820 AJVK01023821 BT127417 AEE62379.1 KB632278 ERL91265.1 APGK01035397 KB740924 ENN78065.1 ABLF02029434 GEMB01002306 JAS00875.1 GFXV01008107 MBW19912.1 GDHC01001769 JAQ16860.1
KQ459053 BAM18121.1 KPJ04189.1 NWSH01003461 PCG66253.1 ODYU01003385 SOQ42077.1 RSAL01000495 RVE41533.1 AGBW02010605 OWR48187.1 MF319745 ASJ26459.1 JTDY01003379 KOB69646.1 LADJ01000094 KPJ20830.1 GDAI01000149 JAI17454.1 CH964272 EDW84673.1 GECU01026713 JAS80993.1 CH902617 EDV42581.1 CH940650 EDW67324.1 JRES01000955 KNC26814.1 CH933806 EDW16311.1 GFDG01001973 JAV16826.1 GFDG01001974 JAV16825.1 CVRI01000047 CRK98059.1 CP012526 ALC47400.1 CM000364 EDX13867.1 CH480815 EDW43129.1 CH916373 EDV94406.1 CH954182 EDV54202.1 GAKP01008179 GAKP01008178 GAKP01008177 GAKP01008175 GAKP01008173 JAC50777.1 CM000070 EAL28118.1 DS232140 EDS35924.1 AE014297 AAN13899.1 AY051764 Y10911 GDHF01031881 GDHF01027384 GDHF01021211 GDHF01013561 GDHF01011070 GDHF01001202 JAI20433.1 JAI24930.1 JAI31103.1 JAI38753.1 JAI41244.1 JAI51112.1 GEBQ01014124 JAT25853.1 CM000160 EDW97945.1 AXCM01004806 OUUW01000005 SPP81057.1 CH479179 EDW24204.1 CCAG010001866 EZ422436 ADD18712.1 KK852902 KDR14073.1 GGFM01001520 MBW22271.1 AAAB01008816 EAA05289.4 NEVH01013262 PNF29067.1 GGFJ01006167 MBW55308.1 KA646141 AFP60770.1 GGFM01006731 MBW27482.1 GAMC01005003 JAC01553.1 GFDL01008137 JAV26908.1 ADMH02000909 ETN64695.1 GAMD01002888 JAA98702.1 APCN01001478 JXJN01005495 KQ971338 EFA02878.1 AXCN02001731 GGFK01003331 MBW36652.1 GBBI01003389 JAC15323.1 ATLV01024337 KE525351 KFB51249.1 GFTR01005827 JAW10599.1 GFDF01001863 JAV12221.1 GANO01001834 JAB58037.1 GAPW01001785 JAC11813.1 GALX01002921 JAB65545.1 JXUM01151271 KQ571179 KXJ68229.1 DQ440338 CH477951 ABF18371.1 EAT34841.1 GECZ01015530 JAS54239.1 AJVK01032324 GEDC01012329 JAS24969.1 QCYY01003201 ROT64718.1 LJIG01016233 KRT81217.1 ACPB03011673 AJWK01023776 AJVK01023820 AJVK01023821 BT127417 AEE62379.1 KB632278 ERL91265.1 APGK01035397 KB740924 ENN78065.1 ABLF02029434 GEMB01002306 JAS00875.1 GFXV01008107 MBW19912.1 GDHC01001769 JAQ16860.1
Proteomes
UP000005204
UP000053268
UP000218220
UP000283053
UP000007151
UP000037510
+ More
UP000053240 UP000007798 UP000007801 UP000192221 UP000008792 UP000037069 UP000009192 UP000183832 UP000092553 UP000000304 UP000001292 UP000001070 UP000008711 UP000001819 UP000002320 UP000000803 UP000002282 UP000095300 UP000075883 UP000268350 UP000008744 UP000075920 UP000076408 UP000092444 UP000027135 UP000007062 UP000075902 UP000235965 UP000095301 UP000076407 UP000000673 UP000092443 UP000075903 UP000075840 UP000092460 UP000069272 UP000075882 UP000007266 UP000075885 UP000075886 UP000075901 UP000075884 UP000092445 UP000075900 UP000030765 UP000075881 UP000078200 UP000069940 UP000249989 UP000008820 UP000075880 UP000092462 UP000091820 UP000283509 UP000015103 UP000092461 UP000030742 UP000019118 UP000007819 UP000192223
UP000053240 UP000007798 UP000007801 UP000192221 UP000008792 UP000037069 UP000009192 UP000183832 UP000092553 UP000000304 UP000001292 UP000001070 UP000008711 UP000001819 UP000002320 UP000000803 UP000002282 UP000095300 UP000075883 UP000268350 UP000008744 UP000075920 UP000076408 UP000092444 UP000027135 UP000007062 UP000075902 UP000235965 UP000095301 UP000076407 UP000000673 UP000092443 UP000075903 UP000075840 UP000092460 UP000069272 UP000075882 UP000007266 UP000075885 UP000075886 UP000075901 UP000075884 UP000092445 UP000075900 UP000030765 UP000075881 UP000078200 UP000069940 UP000249989 UP000008820 UP000075880 UP000092462 UP000091820 UP000283509 UP000015103 UP000092461 UP000030742 UP000019118 UP000007819 UP000192223
Interpro
Gene 3D
CDD
ProteinModelPortal
H9JWZ4
A0A2W1BFM8
S4PPT2
I4DJN1
A0A2A4J4B3
A0A2H1VMP4
+ More
A0A3S2LPC2 A0A212F375 A0A220K8N4 A0A0L7L2W1 A0A0N1IKH2 A0A0K8TSV4 B4NHR3 A0A1B6I241 B3LVL1 A0A1W4VQ24 B4M0N2 A0A0L0C5X1 B4K6L1 A0A1L8EDU1 A0A1L8EDU6 A0A1J1ICX8 A0A0M4F600 B4QZD1 B4HM81 B4JRL6 B3P7P1 A0A034W901 Q297T7 B0WW92 A4V383 P91929 A0A0K8VIR8 A0A1B6LQC9 B4PLJ6 A0A1I8Q2W2 A0A182M9V2 A0A3B0JFE9 B4G309 A0A182WDL5 A0A182YAZ9 D3TLY4 A0A067R4C9 A0A2M3Z1B4 Q7QHC7 A0A182TDY3 A0A2J7QKF4 A0A2M4BRB2 T1PBJ6 A0A2M3ZG26 W8BR54 A0A1Q3FH78 A0A1I8MID9 A0A182X5Q2 W5JP72 A0A1A9YQF0 A0A182VAU3 T1E8C6 A0A182HW95 A0A1B0AY20 A0A182FW35 A0A182KL06 D1ZZE8 A0A182NZQ0 A0A182Q7B1 A0A2M4A7D5 A0A182T4L6 A0A182N286 A0A1B0A1A9 A0A182RW40 A0A023F1W2 A0A084WM05 A0A182JX01 A0A224XRJ6 A0A1L8E0J4 A0A1A9VWM7 U5EVK8 A0A023ERK6 V5G650 A0A182G813 Q1HQV6 A0A182JDF2 A0A1B6FVR3 A0A1B0D7X3 A0A1A9WWW0 A0A1B6DH01 A0A3R7SKG8 A0A0T6B1G8 T1HTY7 A0A1B0CQN1 A0A1B0D3Z1 J3JVZ9 U4UML3 N6U8T6 J9K3B7 A0A170ZEA3 A0A2H8U1Y7 A0A146ME71 A0A1W4XC65
A0A3S2LPC2 A0A212F375 A0A220K8N4 A0A0L7L2W1 A0A0N1IKH2 A0A0K8TSV4 B4NHR3 A0A1B6I241 B3LVL1 A0A1W4VQ24 B4M0N2 A0A0L0C5X1 B4K6L1 A0A1L8EDU1 A0A1L8EDU6 A0A1J1ICX8 A0A0M4F600 B4QZD1 B4HM81 B4JRL6 B3P7P1 A0A034W901 Q297T7 B0WW92 A4V383 P91929 A0A0K8VIR8 A0A1B6LQC9 B4PLJ6 A0A1I8Q2W2 A0A182M9V2 A0A3B0JFE9 B4G309 A0A182WDL5 A0A182YAZ9 D3TLY4 A0A067R4C9 A0A2M3Z1B4 Q7QHC7 A0A182TDY3 A0A2J7QKF4 A0A2M4BRB2 T1PBJ6 A0A2M3ZG26 W8BR54 A0A1Q3FH78 A0A1I8MID9 A0A182X5Q2 W5JP72 A0A1A9YQF0 A0A182VAU3 T1E8C6 A0A182HW95 A0A1B0AY20 A0A182FW35 A0A182KL06 D1ZZE8 A0A182NZQ0 A0A182Q7B1 A0A2M4A7D5 A0A182T4L6 A0A182N286 A0A1B0A1A9 A0A182RW40 A0A023F1W2 A0A084WM05 A0A182JX01 A0A224XRJ6 A0A1L8E0J4 A0A1A9VWM7 U5EVK8 A0A023ERK6 V5G650 A0A182G813 Q1HQV6 A0A182JDF2 A0A1B6FVR3 A0A1B0D7X3 A0A1A9WWW0 A0A1B6DH01 A0A3R7SKG8 A0A0T6B1G8 T1HTY7 A0A1B0CQN1 A0A1B0D3Z1 J3JVZ9 U4UML3 N6U8T6 J9K3B7 A0A170ZEA3 A0A2H8U1Y7 A0A146ME71 A0A1W4XC65
PDB
5LNK
E-value=1.12432e-41,
Score=427
Ontologies
PATHWAY
GO
PANTHER
Topology
Subcellular location
Mitochondrion matrix
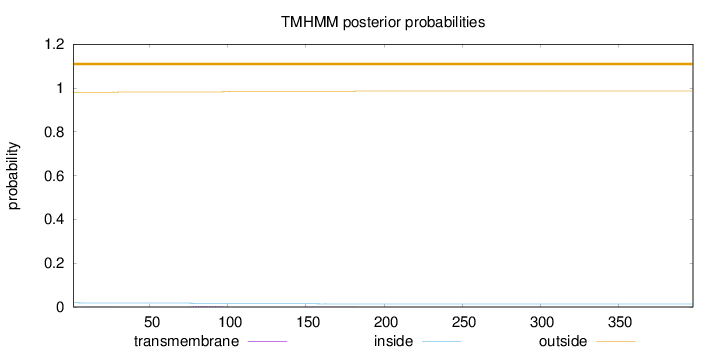
Length:
398
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.10427
Exp number, first 60 AAs:
0.01523
Total prob of N-in:
0.01888
outside
1 - 398
Population Genetic Test Statistics
Pi
179.250167
Theta
141.394781
Tajima's D
1.290341
CLR
0
CSRT
0.739113044347783
Interpretation
Uncertain
