Gene
KWMTBOMO12160
Pre Gene Modal
BGIBMGA004213
Annotation
RNA-directed_RNA_polymerase_L_[Papilio_machaon]
Full name
Large structural protein
+ More
RNA-directed RNA polymerase L
RNA-directed RNA polymerase L
Alternative Name
RNA-directed RNA polymerase L
Large structural protein
Replicase
Transcriptase
Large structural protein
Replicase
Transcriptase
Location in the cell
Nuclear Reliability : 2.97
Sequence
CDS
ATGGTGAATCCGAGTAAAAGACGTAGAGGAAATAATAAAGCTGAAAACTACGAAAAGTTGGTTGAGGATATACTTACAAATTTTAAGGCCATGGGCTGCAGGATGTCATTAAAAGTACATATGCTGCATGCTCATTTGGATAAATTTAAAAACAATATGGGAGCCTATTTCGAAGAGCAAGGAGAACGTTTCCATCAGGACATCATGAATTTTGAACAACGCTATCAAGGCCAATACAATGAAAACATGATGGGCGACTATATTTGGGGTTTATTGAGAGAAAGTAGCTATGAACATAAAAGAAAAAAGCAAACTATGACTAACACTGAGCTGGAAGTGGCTAAAAGAATGGATAAGGTAGCCTCTAACATAAAAGACCAAGGCCACGCCAACTTGAACCTTGATTTGAGTTCTTGGAATCAGCTGAATAGACACGAACTCAATAAGTACCTTTTCCAAGAACTCGACAATCTTCATGGGAAGAAGAACCTCTACTCAGATTCACATTTATGGTATAACCGGTGCCTGGTACTTCTGAGCTCTCGATTAACTCCTCCAAAGATAGGACCAGACGGCATGAGGCAGAAAGCTTGGACCCTCACAACAATAATGATAATAAAAATAGCTCTAGAAGAATGTAACATCCAAGGAGAGATTATGGGACAAGGAGATAACCAGGTTGTGCATATAAAACTTGACAAGGAACAACAGAGTCGGCCCCACTATTACATCAAGCTTCTATTAAACAGTCTAGATTATCTCTTCAGGGCTGCAGGGCTATCTTTGAAATTGCAAGAAACTTGGTATTCACAGAACCTTTTTGAATATTCAAAAGTTCGTTATTATAAATCTATCAGAATAGACGATAGCCTCAAGAGAATAAACAGGATGATACCTGATATTAATGAAGGCTTCCCCTCTTTACAATCCTTAATAACTGGGGTCTCCACCACTACAGAAAACATGTCAAGGAATTACCTGTCCCCAGTCATCCCATTCTTCTTGTATTCTTTAGAACTTGGTAATACTTTAAGACGCAAGAGGATCCTTAACTTAAAACTTCGAGAAACCACAAAATTGTGCGCATTGATGAACATACCATCCATTCTAGGAGGACTTCCCTTATCTAATATGTATCAGCATTGCCAAAGAGGATGCCCTGATCCACTAACTATATGGCTCAAAGTCATTGGGGTTATAAAGAGACAAAGCCCACTGGTTTACAATTCAATTCTCCACCTTATTCCGTGCAGAACGAAGGATCTATATGATCCTGTAAAACTCGTAGAAGATATCTACAGCCTGAATATAGTAGGAATGCCCAATTTCGATAGGAAGGCACGAGAAATTATCGAGGAGTTCCTTCCACAGTATGTAACAAACCCAAAAGTTATTCGGTTGTTGGAAGCTGATAGGATGATAAACGAGGATCCTAGTAGAGAGGAAACAATATTCTCTCTCGGCAAACAAAAAGACGCTGAAGCTGTAGAGGTGCTCAAAAAAAGATTGAAAGGATCTGAATTACCAGTGGTAGGAGATCACTTATTAGAACGCGCCCTTGCCGAGCTCCCGTGCACGTTCAAAGGAGCTCAATGGTTGAGAGAGATAACTTGGGGTTTTCCTATAACTGGAATAACGAGTCCAGTACCTTGTGAGCAAGTTAAAATCCAGGATTTTGATCTCACAGAACCACAAAAGAAACGGGATTGTATCGTAGTAAAAACCTCAATCAAGTTAAAAACAGAGGGTAAATTAGCGCTAAAGAAGAGAGGCCCTTTTGAACCATACTTAGGATCTTCTACTCCGGAGAAAATGATCAAACCTAAACTAACTGTGGTAAACCCTAATCCCCAAGTTAAATCAGTAATGAAACTCTATTATATCCTAACATATCTGGAAAGAATGGATCCTACTAGTGATCTAATTAGCCTGATATAG
Protein
MVNPSKRRRGNNKAENYEKLVEDILTNFKAMGCRMSLKVHMLHAHLDKFKNNMGAYFEEQGERFHQDIMNFEQRYQGQYNENMMGDYIWGLLRESSYEHKRKKQTMTNTELEVAKRMDKVASNIKDQGHANLNLDLSSWNQLNRHELNKYLFQELDNLHGKKNLYSDSHLWYNRCLVLLSSRLTPPKIGPDGMRQKAWTLTTIMIIKIALEECNIQGEIMGQGDNQVVHIKLDKEQQSRPHYYIKLLLNSLDYLFRAAGLSLKLQETWYSQNLFEYSKVRYYKSIRIDDSLKRINRMIPDINEGFPSLQSLITGVSTTTENMSRNYLSPVIPFFLYSLELGNTLRRKRILNLKLRETTKLCALMNIPSILGGLPLSNMYQHCQRGCPDPLTIWLKVIGVIKRQSPLVYNSILHLIPCRTKDLYDPVKLVEDIYSLNIVGMPNFDRKAREIIEEFLPQYVTNPKVIRLLEADRMINEDPSREETIFSLGKQKDAEAVEVLKKRLKGSELPVVGDHLLERALAELPCTFKGAQWLREITWGFPITGITSPVPCEQVKIQDFDLTEPQKKRDCIVVKTSIKLKTEGKLALKKRGPFEPYLGSSTPEKMIKPKLTVVNPNPQVKSVMKLYYILTYLERMDPTSDLISLI
Summary
Description
RNA-directed RNA polymerase that catalyzes the transcription of viral mRNAs, their capping and polyadenylation. The template is composed of the viral RNA tightly encapsidated by the nucleoprotein (N). The viral polymerase binds to the genomic RNA at the 3' leader promoter, and transcribes subsequently all viral mRNAs with a decreasing efficiency. The first gene is the most transcribed, and the last the least transcribed. The viral phosphoprotein acts as a processivity factor. Capping is concommitant with initiation of mRNA transcription. Indeed, a GDP polyribonucleotidyl transferase (PRNTase) adds the cap structure when the nascent RNA chain length has reached few nucleotides. Ribose 2'-O methylation of viral mRNA cap precedes and facilitates subsequent guanine-N-7 methylation, both activities being carried by the viral polymerase. Polyadenylation of mRNAs occur by a stuttering mechanism at a slipery stop site present at the end viral genes. After finishing transcription of a mRNA, the polymerase can resume transcription of the downstream gene.
Displays RNA-directed RNA polymerase activity. The viral mRNA guanylyl transferase displays a different biochemical reaction than the cellular enzyme. The template is composed of the viral RNA tightly encapsidated by the nucleoprotein (N). Functions either as transcriptase or as replicase. The transcriptase synthesizes subsequently three subgenomic RNAs, assuring their capping and polyadenylation by a stuttering mechanism. In replicase mode, the polymerase replicates the whole viral genome without recognizing the transcriptional signals.
Displays RNA-directed RNA polymerase activity. The viral mRNA guanylyl transferase displays a different biochemical reaction than the cellular enzyme. The template is composed of the viral RNA tightly encapsidated by the nucleoprotein (N). Functions either as transcriptase or as replicase. The transcriptase synthesizes subsequently three subgenomic RNAs, assuring their capping and polyadenylation by a stuttering mechanism. In replicase mode, the polymerase replicates the whole viral genome without recognizing the transcriptional signals.
Catalytic Activity
a ribonucleoside 5'-triphosphate + RNA(n) = diphosphate + RNA(n+1)
a 5'-(5'-triphosphoguanosine)-(purine-ribonucleotide)-[mRNA] + S-adenosyl-L-methionine = a 5'-(N(7)-methyl 5'-triphosphoguanosine)-(purine-ribonucleotide)-[mRNA] + S-adenosyl-L-homocysteine
a 5'-(N(7)-methyl 5'-triphosphoguanosine)-(2'-O-methyl-purine-ribonucleotide)-(ribonucleotide)-[mRNA] + S-adenosyl-L-methionine = a 5'-(N(7)-methyl 5'-triphosphoguanosine)-(2'-O-methyl-purine-ribonucleotide)-(2'-O-methyl-ribonucleotide)-[mRNA] + H(+) + S-adenosyl-L-homocysteine
a 5'-triphospho-(purine-ribonucleotide)-[mRNA] + GDP + H(+) = a 5'-(5'-triphosphoguanosine)-(purine-ribonucleotide)-[mRNA] + diphosphate
a 5'-(5'-triphosphoguanosine)-(purine-ribonucleotide)-[mRNA] + S-adenosyl-L-methionine = a 5'-(N(7)-methyl 5'-triphosphoguanosine)-(purine-ribonucleotide)-[mRNA] + S-adenosyl-L-homocysteine
a 5'-(N(7)-methyl 5'-triphosphoguanosine)-(2'-O-methyl-purine-ribonucleotide)-(ribonucleotide)-[mRNA] + S-adenosyl-L-methionine = a 5'-(N(7)-methyl 5'-triphosphoguanosine)-(2'-O-methyl-purine-ribonucleotide)-(2'-O-methyl-ribonucleotide)-[mRNA] + H(+) + S-adenosyl-L-homocysteine
a 5'-triphospho-(purine-ribonucleotide)-[mRNA] + GDP + H(+) = a 5'-(5'-triphosphoguanosine)-(purine-ribonucleotide)-[mRNA] + diphosphate
Subunit
Interacts with the P protein.
Similarity
Belongs to the paramyxovirus L protein family.
Keywords
Alternative splicing
ATP-binding
Complete proteome
Host nucleus
Methyltransferase
mRNA capping
mRNA processing
Multifunctional enzyme
Nucleotide-binding
Nucleotidyltransferase
RNA-directed RNA polymerase
S-adenosyl-L-methionine
Transferase
Viral RNA replication
Virion
Feature
chain RNA-directed RNA polymerase L
Uniprot
H9J3X4
A0A191T7V7
H9J143
A0A0N1PIT2
A0A1L3KN31
A0A0B5KK48
+ More
W4VRB8 A0A140D8N6 A0A1Z2RT94 A0A1Z2RT53 A0A1Z2RSZ4 A0A1Z2RTF5 A0A2Z4HFG4 A0A2H4YI41 A0A2S1ZPS1 A0A2Z4HFJ7 A0A2Z4HFC7 A0A2Z4HFJ1 A0A2Z4HFC6 A0A1Z2RSW5 A0A2Z4HFA6 A0A2Z4GIN2 A0A2Z4HFB8 A0A2Z4HFC4 A0A2Z4HFG9 A0A2Z4HF69 A0A2Z4HF97 A0A2Z4HFS7 A0A1L3KN20 A0A2Z4HF79 A0A0B5KTS7 A0A1C9U5C4 A0A1L3KN72 A0A1Z2RTE0 A0A1Z2RSU8 A0A1Z2RT41 A0A1Z2RT54 A0A1Z2RTB4 A0A3B1EJ88 A0A1L3KML0 A0A3G5FMI8 A0A0J7KAM0 A0A2P1CXU9 A0A2P1CXW0 A0A1B1FIU1 A0A1L3KMW2 A0A1L3KN08 A0A1L3KN23 C4NFK9 C4NFL5 A0A1L3KMN2 A0A0J7KGS3 A0A3B1EJ87 A0A0B5KXM0 M9V5F4 I6Z0N8 A0A0M5I220 M9V8P5 A0A2K9YNH2 B4ZYK0 A0A077EMR8 E7D0B7 I6ZL17 F6JSK0 A0A1B3IT90 C5I0W4 U5L3B0 A0A067YEZ6 A0A0C5KQX4 M9V9S5 G8FRZ0 Q912Y8 A0A2P1GNH0 C7AXL4 I6ZL26 F6JSK6 G9F7N3 A0A1D3JBB3 A2PYK9 A2PYQ5 A0A1Y0BW79 Q912Y9 A0A220SY85 Q912Z3 A0A1D3JBE5 A0A1Y0BW98 Q8JMN0
W4VRB8 A0A140D8N6 A0A1Z2RT94 A0A1Z2RT53 A0A1Z2RSZ4 A0A1Z2RTF5 A0A2Z4HFG4 A0A2H4YI41 A0A2S1ZPS1 A0A2Z4HFJ7 A0A2Z4HFC7 A0A2Z4HFJ1 A0A2Z4HFC6 A0A1Z2RSW5 A0A2Z4HFA6 A0A2Z4GIN2 A0A2Z4HFB8 A0A2Z4HFC4 A0A2Z4HFG9 A0A2Z4HF69 A0A2Z4HF97 A0A2Z4HFS7 A0A1L3KN20 A0A2Z4HF79 A0A0B5KTS7 A0A1C9U5C4 A0A1L3KN72 A0A1Z2RTE0 A0A1Z2RSU8 A0A1Z2RT41 A0A1Z2RT54 A0A1Z2RTB4 A0A3B1EJ88 A0A1L3KML0 A0A3G5FMI8 A0A0J7KAM0 A0A2P1CXU9 A0A2P1CXW0 A0A1B1FIU1 A0A1L3KMW2 A0A1L3KN08 A0A1L3KN23 C4NFK9 C4NFL5 A0A1L3KMN2 A0A0J7KGS3 A0A3B1EJ87 A0A0B5KXM0 M9V5F4 I6Z0N8 A0A0M5I220 M9V8P5 A0A2K9YNH2 B4ZYK0 A0A077EMR8 E7D0B7 I6ZL17 F6JSK0 A0A1B3IT90 C5I0W4 U5L3B0 A0A067YEZ6 A0A0C5KQX4 M9V9S5 G8FRZ0 Q912Y8 A0A2P1GNH0 C7AXL4 I6ZL26 F6JSK6 G9F7N3 A0A1D3JBB3 A2PYK9 A2PYQ5 A0A1Y0BW79 Q912Y9 A0A220SY85 Q912Z3 A0A1D3JBE5 A0A1Y0BW98 Q8JMN0
EC Number
2.7.7.48
Pubmed
EMBL
BABH01041330
KU865643
ANI26478.1
BABH01031416
KQ460794
KPJ12187.1
+ More
KX884430 APG78739.1 KM817638 AJG39135.1 GANO01004103 JAB55768.1 KR822819 AMK09260.1 MF176318 ASA47413.1 MF176298 ASA47379.1 MF176268 ASA47322.1 MF176332 MF176377 ASA47433.1 ASA47504.1 MH430648 MH430659 AWW13443.1 AWW13492.1 MG012486 AUE23903.1 MH237595 AWK27462.1 MH430650 AWW13450.1 MH430656 AWW13482.1 MH430665 AWW13503.1 MH430657 AWW13485.1 MF176247 ASA47292.1 MH430663 MH430664 AWW13497.1 AWW13500.1 MH037149 AWW01093.1 MH430654 AWW13472.1 MH430666 AWW13507.1 MH430651 AWW13453.1 MH430655 AWW13479.1 MH430653 AWW13465.1 MH430652 AWW13458.1 KX884428 APG78729.1 MH430658 AWW13489.1 KM817661 AJG39227.1 KX148553 AOR51378.1 KX884463 APG78861.1 MF176375 ASA47494.1 MF176245 ASA47282.1 MF176296 ASA47369.1 MF176316 ASA47403.1 MF176356 ASA47464.1 MF287670 AWI42881.1 KX884408 APG78650.1 MH477287 AYW51538.1 LBMM01010437 KMQ87458.1 MG550265 MG550266 AVK42865.1 AVK42870.1 MG550267 AVK42875.1 KX257488 ANQ45640.1 KX884407 APG78645.1 KX884436 APG78764.1 KX884441 APG78782.1 FJ554525 ACQ94979.1 FJ554526 ACQ94985.1 KX884406 APG78642.1 LBMM01007576 KMQ89613.1 MF287669 AWI42880.1 KM817649 AJG39173.1 KC595273 AGJ74924.2 JX065210 AFN70809.2 KP972428 ALB34720.1 KC464478 AGJ74903.1 MF360791 AUW34395.1 EU781967 ACG59353.1 KM114265 AIL50416.1 HM998710 ADU05398.1 JX065207 AFN70793.1 GU249595 AEG78314.1 KU748788 AOF44310.1 FJ620690 ACS32310.1 KF578398 AGW46941.1 KF530058 AHA90827.1 KM213865 AJP67524.1 KC464471 AGJ74872.1 JN035149 AER30498.1 AJ311524 CAC70659.1 MG599939 AVM87541.1 FJ665628 ACT21686.1 JN014950 JX065209 AEW69859.1 AFN70803.1 GU249596 AEG41944.1 JN014949 AEW69871.1 LT594381 SBT82866.1 AB246670 BAF46117.1 AB258389 BAF46124.1 KY488724 ART67025.1 AJ311523 CAC70652.1 KY002076 ASK38755.1 AJ311522 CAC70645.1 LT594387 SBT82897.1 KY488727 ART67043.1 AY114161 L27077
KX884430 APG78739.1 KM817638 AJG39135.1 GANO01004103 JAB55768.1 KR822819 AMK09260.1 MF176318 ASA47413.1 MF176298 ASA47379.1 MF176268 ASA47322.1 MF176332 MF176377 ASA47433.1 ASA47504.1 MH430648 MH430659 AWW13443.1 AWW13492.1 MG012486 AUE23903.1 MH237595 AWK27462.1 MH430650 AWW13450.1 MH430656 AWW13482.1 MH430665 AWW13503.1 MH430657 AWW13485.1 MF176247 ASA47292.1 MH430663 MH430664 AWW13497.1 AWW13500.1 MH037149 AWW01093.1 MH430654 AWW13472.1 MH430666 AWW13507.1 MH430651 AWW13453.1 MH430655 AWW13479.1 MH430653 AWW13465.1 MH430652 AWW13458.1 KX884428 APG78729.1 MH430658 AWW13489.1 KM817661 AJG39227.1 KX148553 AOR51378.1 KX884463 APG78861.1 MF176375 ASA47494.1 MF176245 ASA47282.1 MF176296 ASA47369.1 MF176316 ASA47403.1 MF176356 ASA47464.1 MF287670 AWI42881.1 KX884408 APG78650.1 MH477287 AYW51538.1 LBMM01010437 KMQ87458.1 MG550265 MG550266 AVK42865.1 AVK42870.1 MG550267 AVK42875.1 KX257488 ANQ45640.1 KX884407 APG78645.1 KX884436 APG78764.1 KX884441 APG78782.1 FJ554525 ACQ94979.1 FJ554526 ACQ94985.1 KX884406 APG78642.1 LBMM01007576 KMQ89613.1 MF287669 AWI42880.1 KM817649 AJG39173.1 KC595273 AGJ74924.2 JX065210 AFN70809.2 KP972428 ALB34720.1 KC464478 AGJ74903.1 MF360791 AUW34395.1 EU781967 ACG59353.1 KM114265 AIL50416.1 HM998710 ADU05398.1 JX065207 AFN70793.1 GU249595 AEG78314.1 KU748788 AOF44310.1 FJ620690 ACS32310.1 KF578398 AGW46941.1 KF530058 AHA90827.1 KM213865 AJP67524.1 KC464471 AGJ74872.1 JN035149 AER30498.1 AJ311524 CAC70659.1 MG599939 AVM87541.1 FJ665628 ACT21686.1 JN014950 JX065209 AEW69859.1 AFN70803.1 GU249596 AEG41944.1 JN014949 AEW69871.1 LT594381 SBT82866.1 AB246670 BAF46117.1 AB258389 BAF46124.1 KY488724 ART67025.1 AJ311523 CAC70652.1 KY002076 ASK38755.1 AJ311522 CAC70645.1 LT594387 SBT82897.1 KY488727 ART67043.1 AY114161 L27077
Proteomes
UP000005204
UP000053240
UP000202789
UP000270875
UP000270144
UP000280288
+ More
UP000204232 UP000275001 UP000270635 UP000280841 UP000272427 UP000201180 UP000218283 UP000207626 UP000279435 UP000036403 UP000278361 UP000201094 UP000202239 UP000202564 UP000119000 UP000029765 UP000202142 UP000203949 UP000203892 UP000217508 UP000159341 UP000218105 UP000145844 UP000156487 UP000217769 UP000232848 UP000146071 UP000112750 UP000127615 UP000164342 UP000104048 UP000097759 UP000129550 UP000115432 UP000201420 UP000113669 UP000099641 UP000124158 UP000142209 UP000272289 UP000117149 UP000185272
UP000204232 UP000275001 UP000270635 UP000280841 UP000272427 UP000201180 UP000218283 UP000207626 UP000279435 UP000036403 UP000278361 UP000201094 UP000202239 UP000202564 UP000119000 UP000029765 UP000202142 UP000203949 UP000203892 UP000217508 UP000159341 UP000218105 UP000145844 UP000156487 UP000217769 UP000232848 UP000146071 UP000112750 UP000127615 UP000164342 UP000104048 UP000097759 UP000129550 UP000115432 UP000201420 UP000113669 UP000099641 UP000124158 UP000142209 UP000272289 UP000117149 UP000185272
Interpro
ProteinModelPortal
H9J3X4
A0A191T7V7
H9J143
A0A0N1PIT2
A0A1L3KN31
A0A0B5KK48
+ More
W4VRB8 A0A140D8N6 A0A1Z2RT94 A0A1Z2RT53 A0A1Z2RSZ4 A0A1Z2RTF5 A0A2Z4HFG4 A0A2H4YI41 A0A2S1ZPS1 A0A2Z4HFJ7 A0A2Z4HFC7 A0A2Z4HFJ1 A0A2Z4HFC6 A0A1Z2RSW5 A0A2Z4HFA6 A0A2Z4GIN2 A0A2Z4HFB8 A0A2Z4HFC4 A0A2Z4HFG9 A0A2Z4HF69 A0A2Z4HF97 A0A2Z4HFS7 A0A1L3KN20 A0A2Z4HF79 A0A0B5KTS7 A0A1C9U5C4 A0A1L3KN72 A0A1Z2RTE0 A0A1Z2RSU8 A0A1Z2RT41 A0A1Z2RT54 A0A1Z2RTB4 A0A3B1EJ88 A0A1L3KML0 A0A3G5FMI8 A0A0J7KAM0 A0A2P1CXU9 A0A2P1CXW0 A0A1B1FIU1 A0A1L3KMW2 A0A1L3KN08 A0A1L3KN23 C4NFK9 C4NFL5 A0A1L3KMN2 A0A0J7KGS3 A0A3B1EJ87 A0A0B5KXM0 M9V5F4 I6Z0N8 A0A0M5I220 M9V8P5 A0A2K9YNH2 B4ZYK0 A0A077EMR8 E7D0B7 I6ZL17 F6JSK0 A0A1B3IT90 C5I0W4 U5L3B0 A0A067YEZ6 A0A0C5KQX4 M9V9S5 G8FRZ0 Q912Y8 A0A2P1GNH0 C7AXL4 I6ZL26 F6JSK6 G9F7N3 A0A1D3JBB3 A2PYK9 A2PYQ5 A0A1Y0BW79 Q912Y9 A0A220SY85 Q912Z3 A0A1D3JBE5 A0A1Y0BW98 Q8JMN0
W4VRB8 A0A140D8N6 A0A1Z2RT94 A0A1Z2RT53 A0A1Z2RSZ4 A0A1Z2RTF5 A0A2Z4HFG4 A0A2H4YI41 A0A2S1ZPS1 A0A2Z4HFJ7 A0A2Z4HFC7 A0A2Z4HFJ1 A0A2Z4HFC6 A0A1Z2RSW5 A0A2Z4HFA6 A0A2Z4GIN2 A0A2Z4HFB8 A0A2Z4HFC4 A0A2Z4HFG9 A0A2Z4HF69 A0A2Z4HF97 A0A2Z4HFS7 A0A1L3KN20 A0A2Z4HF79 A0A0B5KTS7 A0A1C9U5C4 A0A1L3KN72 A0A1Z2RTE0 A0A1Z2RSU8 A0A1Z2RT41 A0A1Z2RT54 A0A1Z2RTB4 A0A3B1EJ88 A0A1L3KML0 A0A3G5FMI8 A0A0J7KAM0 A0A2P1CXU9 A0A2P1CXW0 A0A1B1FIU1 A0A1L3KMW2 A0A1L3KN08 A0A1L3KN23 C4NFK9 C4NFL5 A0A1L3KMN2 A0A0J7KGS3 A0A3B1EJ87 A0A0B5KXM0 M9V5F4 I6Z0N8 A0A0M5I220 M9V8P5 A0A2K9YNH2 B4ZYK0 A0A077EMR8 E7D0B7 I6ZL17 F6JSK0 A0A1B3IT90 C5I0W4 U5L3B0 A0A067YEZ6 A0A0C5KQX4 M9V9S5 G8FRZ0 Q912Y8 A0A2P1GNH0 C7AXL4 I6ZL26 F6JSK6 G9F7N3 A0A1D3JBB3 A2PYK9 A2PYQ5 A0A1Y0BW79 Q912Y9 A0A220SY85 Q912Z3 A0A1D3JBE5 A0A1Y0BW98 Q8JMN0
PDB
5A22
E-value=8.31357e-12,
Score=172
Ontologies
GO
Topology
Subcellular location
Virion
Host cytoplasm
Host nucleus
Host cytoplasm
Host nucleus
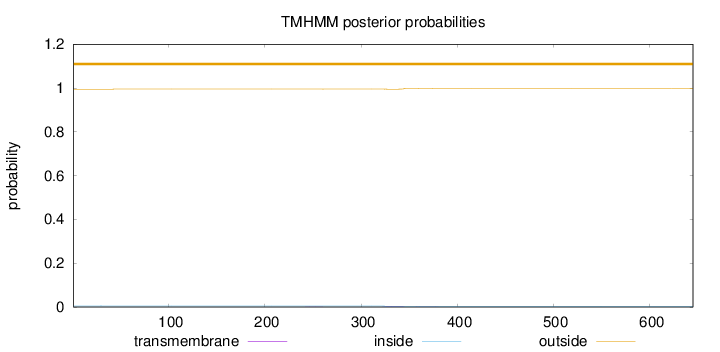
Length:
645
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0757599999999998
Exp number, first 60 AAs:
0.00132
Total prob of N-in:
0.00497
outside
1 - 645
Population Genetic Test Statistics
Pi
209.644966
Theta
206.176494
Tajima's D
0.423499
CLR
0.434678
CSRT
0.494325283735813
Interpretation
Uncertain
