Gene
KWMTBOMO12157
Pre Gene Modal
BGIBMGA008832
Annotation
retrotransposon_protein?_putative?_Ty1-copia_subclass_[Oryza_sativa_Japonica_Group]
Location in the cell
Nuclear Reliability : 2.261
Sequence
CDS
ATGGAAGCCTACATCAATAAAATTAAAGAGTTGGCGTGTCAACTGAGTGAAATGGAATCCGCATTGGATGATGAGTTTATTGCCGTTATAATGCTAAGTGGGCTCACAGAAGATTTTGACCCCCTCATCATGGCAATTGAGAACTCAAATGTAAAACTCACCACGGAACTTGTCTCAGGAAAACTCTTGGCAGAAAACCAGAGGCGTGAGGAGAAACAAGATATCACTGCGTTGGTAACCAAGAAGCCACCGAGATGCTTCAAGTGCAAAAATGTGGGTCATATTTCTAAGAATTGTTCCAACAAAGGAAGCTTCAAGGAGTCAGCGGACGTAGGAGGCAACAAAGGCAAAAATAAATCCAGATATCAGACTAGGAACAATCCGAAGCAGAATATCCCAAAGGACAAAGCGCTCTTTGCAGCTCTGTCTGTAAAAATAAGTAGTGATGTGTGGTACATTGACTCCGGTGCTACTAATCATATGTGCAATGATAAAAAGTACATGATGGACTTCCGACCTAATAGTGTGGATGTGAGCATCGTGAATGGTGAAAAGCTACGAACAGGTGGCATTGGACATGAAGCAATATGTTTATTTTTTGAATACAAAAATGAAACTTTTCGGTATTTCCAACAATTCAAAGCTCTTGTTGAGAAAGAAACGGGAAAGGAATTAAAAATACTTAGGACTGCTAGGGGCGGTGAGTATATGGGTGAGTTTCAGGAATTTCTCAAGAAGAATGGCATCCAACACCAGAAAACTATTGCGGATAGTCCGCAACAGAATGGTGTTGCTGAACGGGCCAATAGGTCCGTCATAGAGAAGGCATGGAGTATGCTGCGGGATCCTAGGCTGGGAATGAAATTTTGGGCTGAGGCTGTGAACACTGCCGTATACCTTAAGAACCGAAGTCCAACCAAAGCTGTGATAGGAGCTACTCCGGAGGAAAAATGGTCCAACAAAGCAAGCAAGAGGGCAAGCTTCGTGAGATTAGTCGACAACCCTGATGATCCCCAGACTGCAGCCGAGGCCTTAAACGGGCCTGAATCGAACAGTTGGAGAAAGGCAATGGCAGATGAGTATAATTCCTTTGTTAAGAACAAGTGTTGGACCCTGACTGAGCTTGAACCCAGCCACAAACCAGTCAAAAGTAAATGGATTTTCAAAAGAAAATATGGAGCAGATGGAGAGTTACTAAAGTATAAAGCTCGTTTAGTGGCCAAGGGTTACACACAGAAGTACGGTATTGATTACCAAGAGACTTTTTCACCAGTGGTAAGGTACTCTACAATTAGGATTTTACTTGCTTTAGCAGCTGAACATGGCCTGTCCATTGAGCACCTGGATGTCAAAATGGTGTTCCTTAATGACGACCTCGAGGAAACTGTATACATGGAGCAGCCAGAGTTCTTTCAAGAAAAAGGAAAAGAGAACATGGTGTACAGGCTAAATAAAGCTGTGTACGGCTTAAAACAGGCATCCAAGACATGGTATGACAAAATTACAAAGATTTTGTGTCACAAACTACTATTCAAGCAATTAAGCACTGAGCCATGTGTCTACTTTAAAAGTGAAAATAAGTCTATGATGATAATAGTTCTCTATGTCGATGACCCATTGTTTTCACATCCCCCAACTTTCAAGGAAAACAGAAAATAA
Protein
MEAYINKIKELACQLSEMESALDDEFIAVIMLSGLTEDFDPLIMAIENSNVKLTTELVSGKLLAENQRREEKQDITALVTKKPPRCFKCKNVGHISKNCSNKGSFKESADVGGNKGKNKSRYQTRNNPKQNIPKDKALFAALSVKISSDVWYIDSGATNHMCNDKKYMMDFRPNSVDVSIVNGEKLRTGGIGHEAICLFFEYKNETFRYFQQFKALVEKETGKELKILRTARGGEYMGEFQEFLKKNGIQHQKTIADSPQQNGVAERANRSVIEKAWSMLRDPRLGMKFWAEAVNTAVYLKNRSPTKAVIGATPEEKWSNKASKRASFVRLVDNPDDPQTAAEALNGPESNSWRKAMADEYNSFVKNKCWTLTELEPSHKPVKSKWIFKRKYGADGELLKYKARLVAKGYTQKYGIDYQETFSPVVRYSTIRILLALAAEHGLSIEHLDVKMVFLNDDLEETVYMEQPEFFQEKGKENMVYRLNKAVYGLKQASKTWYDKITKILCHKLLFKQLSTEPCVYFKSENKSMMIIVLYVDDPLFSHPPTFKENRK
Summary
Uniprot
EMBL
Proteomes
Pfam
Interpro
IPR036397
RNaseH_sf
+ More
IPR039537 Retrotran_Ty1/copia-like
IPR001584 Integrase_cat-core
IPR012337 RNaseH-like_sf
IPR013103 RVT_2
IPR039633 PAP
IPR006843 PAP/fibrillin_dom
IPR025724 GAG-pre-integrase_dom
IPR025314 DUF4219
IPR001878 Znf_CCHC
IPR036875 Znf_CCHC_sf
IPR041588 Integrase_H2C2
IPR002156 RNaseH_domain
IPR000477 RT_dom
IPR021109 Peptidase_aspartic_dom_sf
IPR005162 Retrotrans_gag_dom
IPR041577 RT_RNaseH_2
IPR039537 Retrotran_Ty1/copia-like
IPR001584 Integrase_cat-core
IPR012337 RNaseH-like_sf
IPR013103 RVT_2
IPR039633 PAP
IPR006843 PAP/fibrillin_dom
IPR025724 GAG-pre-integrase_dom
IPR025314 DUF4219
IPR001878 Znf_CCHC
IPR036875 Znf_CCHC_sf
IPR041588 Integrase_H2C2
IPR002156 RNaseH_domain
IPR000477 RT_dom
IPR021109 Peptidase_aspartic_dom_sf
IPR005162 Retrotrans_gag_dom
IPR041577 RT_RNaseH_2
Gene 3D
ProteinModelPortal
Ontologies
KEGG
GO
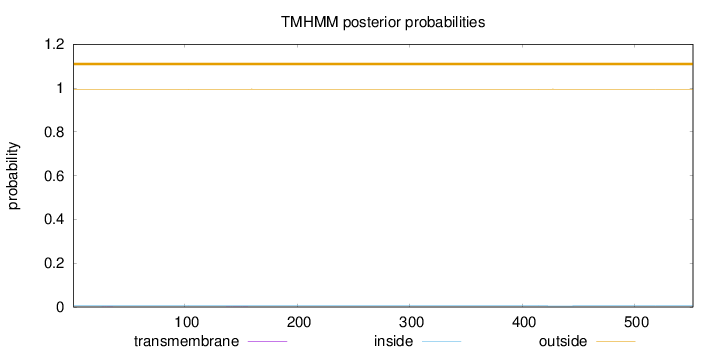
Topology
Length:
552
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00971999999999998
Exp number, first 60 AAs:
0.00263
Total prob of N-in:
0.00547
outside
1 - 552
Population Genetic Test Statistics
Pi
176.270143
Theta
207.554598
Tajima's D
-0.556322
CLR
0.902242
CSRT
0.233288335583221
Interpretation
Uncertain
