Gene
KWMTBOMO12150
Pre Gene Modal
BGIBMGA004258
Annotation
polyprotein_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 2.011 Nuclear Reliability : 1.44
Sequence
CDS
ATGTCTTTAGAAGACAACGAAAGCTTCACAGTTTCACGGGACAACGAGAACTGGTACATAGGTAATGGCGCAACAAGCCACGTCACCAACAGAGCTGACCTTTTCCAAACCTACGAACATTTCAATGAGAACTACACTGTTACAACGGCCGCTGGTAATGAAGTCCGTGCCAAAGGCAAAGGAACAGTACAGCTCAGAGATGTTAACGGGAAACAAATCGACATCAGCCTGCAAGACGTCTGGTACGTTCCGGCCATAAGCAAGAACCTCTTCTCCGTACTCGCACTTCACGATCGAGTCCCGAATAGCACCTTTTCATCCAGGACAACTGCATGCAACGTCATTGTCAACGGGAAAGTATGCTTGATAGGGAAGAGACAAAAACAAGGTGGCTTATATCGACTGGAAGTAAAACCCGTATTGCCAGAAAAACCACCCGAAGTCTTTGCAACTACAGCAACGATGCAACTGTACCACGAGAGACTCGCTCACCAAAATAAAAGACACGTAAGAGCGACGATCCAACGGGAACTAGGTCTAATCCTGCCGATGGATAAAGATACCTGTGAAGGCTGCATCTTTGGAAAGGCGCATGGACTTAAATTCGGTACAAGAACCCGAGCTACCGCGCCCGGAGAAATCGTACACACCGACGTTTGCGGCCCGTTTCAACCTAGCTTCTCAAACTATAAGTACTACGTTCTGTTTAAAGACGACTTTACTGGCTATCGTATGGTGTACTTCATACGCAAGAAGTCCGAGGTCAAAGACAAGTTAGTTTTGATGATCGCTCAAACTAAAACAGTCGGACACACCATAAAATGCCTTCTAAGTGACAACGGCGGTGAATTTGACAACGCGTCCATAAAAAGTGTTTTAGACACACATGGAATAGCACAGCGACTCGTCACCCCGTATACACCTGAACAAAACGGAGTAAGCGTAACGGAGATAGCAAGATCAATGATGTACGCTCATGAGCCATTACCGCAAGTGCTATGGGCAGAGCTCATTAATACCGCCGCATACATACTGAACAGAACAGGTCCCACAAGTTCAGATAAATCTCCGCATGAACTTTGGTATGGAAAGAAACCTATTATTAAACATCTCCGAATAATAGGCTCAATCGCATACGTCCATATTCCAAAGCAGAAGCGTAGAAAATTCGACAAAAAGGCGATAAAAGGTCGACTTATCGGGTACGACGAAGATTGCTACCGTATCTGGATCACAGACGGAAAACACTCGACGCTCGTCCGCTCTCGTGACGTAATCTTCGATGAGAAACCTCTTCCTATCTGTAATAGTGAAATTCCTAAAACAGAAGTTGAGACAAGAGAATACCCGATTACCATAACGCCAAATACGAACTTAGAAAGAGAAGTTACAATTCCAGAGCAAACACACGTTCACGAGAGAGATACAACTACCGAAACAATCTCCAACGAGCATGATCATGTTCTCGAGGGAGAAGAGTCGGATCAAGTCCTCTACGAAAACCATGACCATGAAGACGAAATGACAGTTCCAGAAGAATTTTATGACGCTGAACCACCGCCAACCGACTATAATCTTAGAGATCGCTCACTGATTCGACGTCCTGAGAGACTAAGAGATTACGTCTGTATCACTGAAACATTCCCAAATAATTTCGCAGACGCTATGAAAAGGGACGATCGCGAAGAATGGCAAAGAGCAATGGATAGCGAATTGAAATCCTTCGAAGACCTGAACGTTTGGACGCCGTGTGCATTGCCAAAAGGACGAAAAGCACTTCCCTGCAAATGGTTTTTACGGATCAAAGGAAATCCAGATGGCACGATCGACAAATACAAAGCTCGATTTGTCGTAAAAGGGTACAGACAAAGGAAAGGAGTAGACTATGACCAGACCTTCAGTCCTGTGGCACGCCTGACTACTGTACGAGCACTACTCAGTGTTAGTGCGCATGAAAACTTACATCTGGTCCAATTCGACGTCTGCACAGCCTTTCTGAACGGAAAGCTCGAGGAAGAAATATACGTTCAATCGCCAGAAGGGTGCAACCTCACGGAAGGAACGTCTACCGTTTTCAAGCTTAACCGTAGCCTGTACGGACTTAAACAAGCACCAAGTGGGTATGAATGA
Protein
MSLEDNESFTVSRDNENWYIGNGATSHVTNRADLFQTYEHFNENYTVTTAAGNEVRAKGKGTVQLRDVNGKQIDISLQDVWYVPAISKNLFSVLALHDRVPNSTFSSRTTACNVIVNGKVCLIGKRQKQGGLYRLEVKPVLPEKPPEVFATTATMQLYHERLAHQNKRHVRATIQRELGLILPMDKDTCEGCIFGKAHGLKFGTRTRATAPGEIVHTDVCGPFQPSFSNYKYYVLFKDDFTGYRMVYFIRKKSEVKDKLVLMIAQTKTVGHTIKCLLSDNGGEFDNASIKSVLDTHGIAQRLVTPYTPEQNGVSVTEIARSMMYAHEPLPQVLWAELINTAAYILNRTGPTSSDKSPHELWYGKKPIIKHLRIIGSIAYVHIPKQKRRKFDKKAIKGRLIGYDEDCYRIWITDGKHSTLVRSRDVIFDEKPLPICNSEIPKTEVETREYPITITPNTNLEREVTIPEQTHVHERDTTTETISNEHDHVLEGEESDQVLYENHDHEDEMTVPEEFYDAEPPPTDYNLRDRSLIRRPERLRDYVCITETFPNNFADAMKRDDREEWQRAMDSELKSFEDLNVWTPCALPKGRKALPCKWFLRIKGNPDGTIDKYKARFVVKGYRQRKGVDYDQTFSPVARLTTVRALLSVSAHENLHLVQFDVCTAFLNGKLEEEIYVQSPEGCNLTEGTSTVFKLNRSLYGLKQAPSGYE
Summary
Uniprot
B3Y003
A0A1Y1K1E5
A0A1Y1KNM9
A0A1Y3EJI6
A0A085MW59
A0A0V1NHV8
+ More
A0A0V1M1K0 A0A0V1B5K0 A0A0V1B5A2 A0A0V1K2T2 A0A0A9YL44 A0A1Y3E8D5 A0A0N5E2R3 A0A0V1IPN2 A0A0V1DTZ3 A0A085NBE7 A0A0V0WRK3 A0A085MSN1 A0A085NBL2 J9LCG1 A0A085N6E0 A0A1Y1LDU4 A0A139WFV5 A0A0J7K744 A0A1Y1KR09 A0A1Y1K5W6 A0A1Y1JZX1 A0A1X7U495 X1WMK8 A0A0A9YB53 A0A0A9W6P7 A0A0P6AI73 A0A0J7K9W0 A0A0J7K725 A0A0P5D6G1 A0A0P5AJS8 A0A0P5DII6 A0A2M4CWP1 A0A2M3YZL7 K7JBX1 A0A0J7KRG1 A0A2M4CV35 A0A2M3YZL3 A0A0J7KNZ2 A0A0V0RE76 A0A0J7KBA1 A0A0N7ZJE6 A0A2J7QWT7 A0A182GSW5 A0A2K3NXK0 A0A0J7KM48 A0A1W7R6A3 A0A0J7KDE2 A0A151R7U0 A0A1Y1NHZ0 A0A0V1NED0 A0A0P6B8Y5 A0A2K3MYT4 A0A0P6APU8 A0A1W7R6C3 A0A194RU38 A0A1W7R6J6 A0A2A4JRI1 A0A1W7R6K6 A0A182G1G7 A0A2Z6N0G5 A0A2K3MZ63 A0A0V1L945 A0A1Y1LCN3 A0A1J3CK86 A0A251UIF2 A0A182GA29 A0A2J7Q1Z2 A0A1W7R6D9 H3H839 A0A251T6N3 X1XQX4 A0A0A9YMV4 A0A0N7ZV66 W8BYI7 A0A2J7R668 A0A2J7RKJ6 A0A0P6JS54 A0A0P6IW88 A0A2J7R8S8 B6V6Z8 Q9ZRJ0 J9KJ72 A0A182GZN1 A0A392M193 Q7Y141 A0A1W7R685
A0A0V1M1K0 A0A0V1B5K0 A0A0V1B5A2 A0A0V1K2T2 A0A0A9YL44 A0A1Y3E8D5 A0A0N5E2R3 A0A0V1IPN2 A0A0V1DTZ3 A0A085NBE7 A0A0V0WRK3 A0A085MSN1 A0A085NBL2 J9LCG1 A0A085N6E0 A0A1Y1LDU4 A0A139WFV5 A0A0J7K744 A0A1Y1KR09 A0A1Y1K5W6 A0A1Y1JZX1 A0A1X7U495 X1WMK8 A0A0A9YB53 A0A0A9W6P7 A0A0P6AI73 A0A0J7K9W0 A0A0J7K725 A0A0P5D6G1 A0A0P5AJS8 A0A0P5DII6 A0A2M4CWP1 A0A2M3YZL7 K7JBX1 A0A0J7KRG1 A0A2M4CV35 A0A2M3YZL3 A0A0J7KNZ2 A0A0V0RE76 A0A0J7KBA1 A0A0N7ZJE6 A0A2J7QWT7 A0A182GSW5 A0A2K3NXK0 A0A0J7KM48 A0A1W7R6A3 A0A0J7KDE2 A0A151R7U0 A0A1Y1NHZ0 A0A0V1NED0 A0A0P6B8Y5 A0A2K3MYT4 A0A0P6APU8 A0A1W7R6C3 A0A194RU38 A0A1W7R6J6 A0A2A4JRI1 A0A1W7R6K6 A0A182G1G7 A0A2Z6N0G5 A0A2K3MZ63 A0A0V1L945 A0A1Y1LCN3 A0A1J3CK86 A0A251UIF2 A0A182GA29 A0A2J7Q1Z2 A0A1W7R6D9 H3H839 A0A251T6N3 X1XQX4 A0A0A9YMV4 A0A0N7ZV66 W8BYI7 A0A2J7R668 A0A2J7RKJ6 A0A0P6JS54 A0A0P6IW88 A0A2J7R8S8 B6V6Z8 Q9ZRJ0 J9KJ72 A0A182GZN1 A0A392M193 Q7Y141 A0A1W7R685
Pubmed
EMBL
AB378753
BAG68376.1
GEZM01099107
JAV53485.1
GEZM01078128
GEZM01078127
+ More
JAV63002.1 LVZM01009926 OUC45303.1 KL367622 KFD61455.1 JYDM01000206 KRZ83588.1 JYDO01000312 KRZ65659.1 JYDH01000102 KRY32233.1 KRY32232.1 JYDV01000020 KRZ41499.1 GBHO01013359 JAG30245.1 LVZM01021469 OUC41373.1 JYDV01000213 KRZ24707.1 JYDR01000237 KRY65040.1 KL367520 KFD66793.1 JYDK01000067 KRX78390.1 KL367684 KFD60227.1 KFD66858.1 ABLF02020242 ABLF02020246 ABLF02042026 KL367545 KFD65036.1 GEZM01063217 JAV69187.1 KQ971352 KYB26731.1 LBMM01012672 KMQ86059.1 GEZM01078105 JAV63008.1 GEZM01096605 JAV54776.1 GEZM01096656 JAV54693.1 ABLF02066512 GBHO01016859 JAG26745.1 GBHO01043059 JAG00545.1 GDIP01034138 JAM69577.1 LBMM01010801 KMQ87218.1 LBMM01012536 KMQ86167.1 GDIP01162319 JAJ61083.1 GDIP01198316 JAJ25086.1 GDIP01156971 JAJ66431.1 GGFL01005562 MBW69740.1 GGFM01000952 MBW21703.1 AAZX01004352 LBMM01003976 KMQ92933.1 GGFL01004951 MBW69129.1 GGFM01000953 MBW21704.1 LBMM01004825 KMQ92052.1 JYDL01000266 KRX12804.1 LBMM01010133 KMQ87653.1 GDIP01239569 JAI83832.1 NEVH01009420 PNF33053.1 JXUM01085595 KQ563513 KXJ73717.1 ASHM01002084 PNY07765.1 LBMM01005619 KMQ91367.1 GEHC01000959 JAV46686.1 LBMM01008998 KMQ88473.1 KQ483989 KYP38573.1 GEZM01007166 GEZM01007165 JAV95327.1 JYDM01000301 KRZ82363.1 GDIP01019956 JAM83759.1 ASHM01014005 PNX95963.1 GDIP01039936 JAM63779.1 GEHC01000928 JAV46717.1 KQ459875 KPJ19636.1 GEHC01000868 JAV46777.1 NWSH01000816 PCG74073.1 GEHC01000867 JAV46778.1 JXUM01136904 KQ568499 KXJ68982.1 DF973673 GAU37486.1 ASHM01014140 PNX96091.1 JYDW01000105 KRZ55879.1 GEZM01063091 JAV69316.1 GEVI01023957 JAU08363.1 CM007895 OTG22666.1 JXUM01155348 KQ572178 KXJ68066.1 NEVH01019375 PNF22597.1 GEHC01000923 JAV46722.1 DS567278 CM007900 OTG06758.1 ABLF02042030 GBHO01012744 JAG30860.1 GDIP01209409 JAJ13993.1 GAMC01008189 JAB98366.1 NEVH01006980 PNF36330.1 NEVH01002725 PNF41351.1 GDUN01000696 JAN95223.1 GDUN01000697 JAN95222.1 NEVH01006721 PNF37230.1 FJ238509 ACI62137.1 D83003 BAA11674.1 ABLF02041793 JXUM01099923 KQ564528 KXJ72133.1 LXQA010000987 MCH80374.1 AC133339 DP000009 AAP46257.1 ABF99717.1 GEHC01000966 JAV46679.1
JAV63002.1 LVZM01009926 OUC45303.1 KL367622 KFD61455.1 JYDM01000206 KRZ83588.1 JYDO01000312 KRZ65659.1 JYDH01000102 KRY32233.1 KRY32232.1 JYDV01000020 KRZ41499.1 GBHO01013359 JAG30245.1 LVZM01021469 OUC41373.1 JYDV01000213 KRZ24707.1 JYDR01000237 KRY65040.1 KL367520 KFD66793.1 JYDK01000067 KRX78390.1 KL367684 KFD60227.1 KFD66858.1 ABLF02020242 ABLF02020246 ABLF02042026 KL367545 KFD65036.1 GEZM01063217 JAV69187.1 KQ971352 KYB26731.1 LBMM01012672 KMQ86059.1 GEZM01078105 JAV63008.1 GEZM01096605 JAV54776.1 GEZM01096656 JAV54693.1 ABLF02066512 GBHO01016859 JAG26745.1 GBHO01043059 JAG00545.1 GDIP01034138 JAM69577.1 LBMM01010801 KMQ87218.1 LBMM01012536 KMQ86167.1 GDIP01162319 JAJ61083.1 GDIP01198316 JAJ25086.1 GDIP01156971 JAJ66431.1 GGFL01005562 MBW69740.1 GGFM01000952 MBW21703.1 AAZX01004352 LBMM01003976 KMQ92933.1 GGFL01004951 MBW69129.1 GGFM01000953 MBW21704.1 LBMM01004825 KMQ92052.1 JYDL01000266 KRX12804.1 LBMM01010133 KMQ87653.1 GDIP01239569 JAI83832.1 NEVH01009420 PNF33053.1 JXUM01085595 KQ563513 KXJ73717.1 ASHM01002084 PNY07765.1 LBMM01005619 KMQ91367.1 GEHC01000959 JAV46686.1 LBMM01008998 KMQ88473.1 KQ483989 KYP38573.1 GEZM01007166 GEZM01007165 JAV95327.1 JYDM01000301 KRZ82363.1 GDIP01019956 JAM83759.1 ASHM01014005 PNX95963.1 GDIP01039936 JAM63779.1 GEHC01000928 JAV46717.1 KQ459875 KPJ19636.1 GEHC01000868 JAV46777.1 NWSH01000816 PCG74073.1 GEHC01000867 JAV46778.1 JXUM01136904 KQ568499 KXJ68982.1 DF973673 GAU37486.1 ASHM01014140 PNX96091.1 JYDW01000105 KRZ55879.1 GEZM01063091 JAV69316.1 GEVI01023957 JAU08363.1 CM007895 OTG22666.1 JXUM01155348 KQ572178 KXJ68066.1 NEVH01019375 PNF22597.1 GEHC01000923 JAV46722.1 DS567278 CM007900 OTG06758.1 ABLF02042030 GBHO01012744 JAG30860.1 GDIP01209409 JAJ13993.1 GAMC01008189 JAB98366.1 NEVH01006980 PNF36330.1 NEVH01002725 PNF41351.1 GDUN01000696 JAN95223.1 GDUN01000697 JAN95222.1 NEVH01006721 PNF37230.1 FJ238509 ACI62137.1 D83003 BAA11674.1 ABLF02041793 JXUM01099923 KQ564528 KXJ72133.1 LXQA010000987 MCH80374.1 AC133339 DP000009 AAP46257.1 ABF99717.1 GEHC01000966 JAV46679.1
Proteomes
PRIDE
Interpro
Gene 3D
ProteinModelPortal
B3Y003
A0A1Y1K1E5
A0A1Y1KNM9
A0A1Y3EJI6
A0A085MW59
A0A0V1NHV8
+ More
A0A0V1M1K0 A0A0V1B5K0 A0A0V1B5A2 A0A0V1K2T2 A0A0A9YL44 A0A1Y3E8D5 A0A0N5E2R3 A0A0V1IPN2 A0A0V1DTZ3 A0A085NBE7 A0A0V0WRK3 A0A085MSN1 A0A085NBL2 J9LCG1 A0A085N6E0 A0A1Y1LDU4 A0A139WFV5 A0A0J7K744 A0A1Y1KR09 A0A1Y1K5W6 A0A1Y1JZX1 A0A1X7U495 X1WMK8 A0A0A9YB53 A0A0A9W6P7 A0A0P6AI73 A0A0J7K9W0 A0A0J7K725 A0A0P5D6G1 A0A0P5AJS8 A0A0P5DII6 A0A2M4CWP1 A0A2M3YZL7 K7JBX1 A0A0J7KRG1 A0A2M4CV35 A0A2M3YZL3 A0A0J7KNZ2 A0A0V0RE76 A0A0J7KBA1 A0A0N7ZJE6 A0A2J7QWT7 A0A182GSW5 A0A2K3NXK0 A0A0J7KM48 A0A1W7R6A3 A0A0J7KDE2 A0A151R7U0 A0A1Y1NHZ0 A0A0V1NED0 A0A0P6B8Y5 A0A2K3MYT4 A0A0P6APU8 A0A1W7R6C3 A0A194RU38 A0A1W7R6J6 A0A2A4JRI1 A0A1W7R6K6 A0A182G1G7 A0A2Z6N0G5 A0A2K3MZ63 A0A0V1L945 A0A1Y1LCN3 A0A1J3CK86 A0A251UIF2 A0A182GA29 A0A2J7Q1Z2 A0A1W7R6D9 H3H839 A0A251T6N3 X1XQX4 A0A0A9YMV4 A0A0N7ZV66 W8BYI7 A0A2J7R668 A0A2J7RKJ6 A0A0P6JS54 A0A0P6IW88 A0A2J7R8S8 B6V6Z8 Q9ZRJ0 J9KJ72 A0A182GZN1 A0A392M193 Q7Y141 A0A1W7R685
A0A0V1M1K0 A0A0V1B5K0 A0A0V1B5A2 A0A0V1K2T2 A0A0A9YL44 A0A1Y3E8D5 A0A0N5E2R3 A0A0V1IPN2 A0A0V1DTZ3 A0A085NBE7 A0A0V0WRK3 A0A085MSN1 A0A085NBL2 J9LCG1 A0A085N6E0 A0A1Y1LDU4 A0A139WFV5 A0A0J7K744 A0A1Y1KR09 A0A1Y1K5W6 A0A1Y1JZX1 A0A1X7U495 X1WMK8 A0A0A9YB53 A0A0A9W6P7 A0A0P6AI73 A0A0J7K9W0 A0A0J7K725 A0A0P5D6G1 A0A0P5AJS8 A0A0P5DII6 A0A2M4CWP1 A0A2M3YZL7 K7JBX1 A0A0J7KRG1 A0A2M4CV35 A0A2M3YZL3 A0A0J7KNZ2 A0A0V0RE76 A0A0J7KBA1 A0A0N7ZJE6 A0A2J7QWT7 A0A182GSW5 A0A2K3NXK0 A0A0J7KM48 A0A1W7R6A3 A0A0J7KDE2 A0A151R7U0 A0A1Y1NHZ0 A0A0V1NED0 A0A0P6B8Y5 A0A2K3MYT4 A0A0P6APU8 A0A1W7R6C3 A0A194RU38 A0A1W7R6J6 A0A2A4JRI1 A0A1W7R6K6 A0A182G1G7 A0A2Z6N0G5 A0A2K3MZ63 A0A0V1L945 A0A1Y1LCN3 A0A1J3CK86 A0A251UIF2 A0A182GA29 A0A2J7Q1Z2 A0A1W7R6D9 H3H839 A0A251T6N3 X1XQX4 A0A0A9YMV4 A0A0N7ZV66 W8BYI7 A0A2J7R668 A0A2J7RKJ6 A0A0P6JS54 A0A0P6IW88 A0A2J7R8S8 B6V6Z8 Q9ZRJ0 J9KJ72 A0A182GZN1 A0A392M193 Q7Y141 A0A1W7R685
PDB
3S3O
E-value=0.00101518,
Score=103
Ontologies
GO
PANTHER
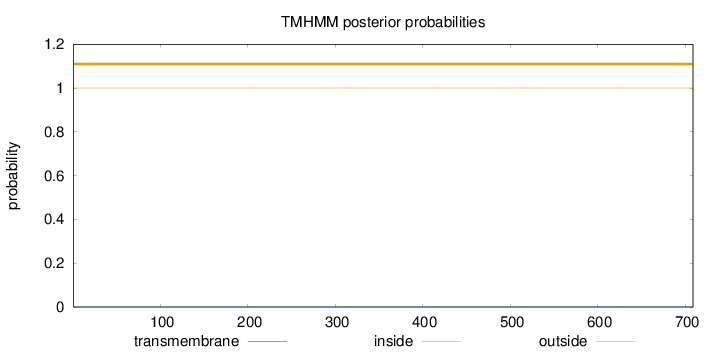
Topology
Length:
709
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0013
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00012
outside
1 - 709
Population Genetic Test Statistics
Pi
0.609524
Theta
3.289592
Tajima's D
-2.150013
CLR
3031.601711
CSRT
0.00499975001249937
Interpretation
Possibly Positive selection
