Gene
KWMTBOMO12142
Annotation
Yokozuna_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 4.208
Sequence
CDS
ATGCATTCCGATACCGAACAAGACTTTTATGCTGTTTCAAGATTCCCACGTGTCCTTGGTACACTTGATGGTACACACATTAGGATTCAGTCACCATGTTCTCAGATTGAAGAAGAATTTAGAAACCGAAAAGGTTACTTTTCCCTTAATGTCCAAGCTGTGTGCAGCAATGCAGATTTATTATTTATGAATGTGGTGGCTCGTTGGCCAGGTTCAGCTCATGATGCCACAATATTTAATAATTCAGACTTGCGAGCTCAGTGTGAAAATGGCTCATTTGGAAATAAGTGGTTACTTGGAGATAGTGCTTATCCCTTGAAGCCATATTTATTAACGCCACTAATTAACACACAGACACGGGGCCAACAGCTTTATAATGAAGCCCATATTCGAACAAGAAACTGCATTGAAAGATGTTTTGGAGTTTGGAAACGAAGATTCCCTGTTGTAGCCTTAACATTGCGCCTGAGCCTTCTAAGAGCGAATGCAGTGATTATAGCTACAGCTGTACTCCACAACATTTGTCGACTGAAGAAGCTTGATGATATATCACCAGAAGTAGAAATTCCTGAGGCAGATGTGTTACCTACAGAAAGTAGCACTGAGGAAACTTTTAAGGAAAAGGAATGCAAAGAAGCAATTGAAGATGGCATAACAGAGGAAATATTGAAGTCAGAAAAATTCAAAAAGAAAGACGCAAAAGCTAGAAGTTTGATAGTCAACTGCCTCTCAGACAAACACTTGGAATACGTCAGAAGCGCTACGAGTGCAAAAGAGATGATCGAAAACTTACGAAAAATATTCAAAAGAAAAAGTACGTTATCTGCGTTATACGTCAGGAAGAAATTATTAACCTTAAAATGTGACCCCAGTGTCGAACTGAGCGACCACTTCAACAATTTTGATATGTTAATAAGACAATTAGAAGAAACTGGAAGCACAGTAAATGAACAAGACAAGGTGTGCCACTTACTACTGACTATGCCTGACAGCTACAATACTACAATTGCCGCCTTAGAAACTACTAATGTGGAACTTACAACAGAGTATGTCAAAAGCAAATTGTTAGATGCAGAACTAAAAAACCAAAATAATTCAATAGAATCACAGGATGCACACAGCTTCCTCAGTTGCTTTAAATGTGGTAAAACTGGTCACAAAGCTATACAGTGCAGAACTGGACACTTAATGAGAGGACAGCATCGAGGCAAAATGGGCAACGTCGGATATTTCAACGGAAGTAACACTAGAGACTTCTATCGAGGCAACGGCAGAGACTTCAATCCAGGTAACGGCAGAGGCTTTAACCGAGGCAATCCCAGAGGCAACGTGAGAGGACAATCGAGGGGATCATCAAGGGACATCATACCCCAACAAGGAAAAAGTGCTTTAAGTGAGAGAAATCCATTTTCTTTTGTTGCCAGTGAAAAGGTACTATCTGTTGATGTGTATAATGAGGACATTAAGTTTATTATTGATTCAGGAGCATCTCAAAATTTAGTCATTGATAGTTATGAAAAGTACATGTCAAACATTGAATATTTAGAAACTAAAACTAAGATTTATGTTGCTAATGGTCAATATTTGTTTTCAAGTAAAAAAGGGATTTTAAATGTAAAATATAATAACTTACCTATTAAAATAGAAGCACTACTTGTCAAGGGGCTATCCCACAACTTATTATCTGTTAAAAAATTATTAGAGAAAGGAAATTCTGTCAGATTTCATAAAAATTCTGTATCAATTGCTAAAGGAAATAACATAATTTATGGTACCATGCTTAATAGTCTGTATGCAATCAATCTTACATTAAACTTAGAAAAATGTTATTCGATAAACAATGATAAAGATCTATGGCATAAAAGGTTAGGACATGCTAATAGAAAACACTTAAAATTATTGAAATTACCTATCTCAGAAAAACCATGTGGAATATGTGTAGAAGGAAAATCAACAAGATTACCATTCTCTACAACACCCAGGCCGAGATCAAAATATATAGGAGAACTCATACACACAGATATATCAGGACCAATAAATATTCCTACACTTACAAATGAGGTATATTTTCATACAATTATAGATGATTACACACATTTCTGTGAGGTATACTTATTACAAAGAAAAAGTGAGGCAACCGATAGGCTCATAGAATATGTTAATAGAATGGAAAGACAAATAGAATGTAAAGTCAAGAAAATCAGAAGTGATAATGGTGGTGAGTTTAAAAATGAAAAACTAAATAAATTTTGCAAAGATAAGGGAATAGTTCAACAGTTTACTTTGCCTTATAGCCCACAGTCAAACGGAGTTTCAGAACGCATGAACAGAAATATTTATAACAGAGCTCGAACTTTATTAATTGAATCTGGTTTACCAAAAACCTTATGGGGTGAAGCGGTGCGATGTGCTGTCTATCAAACCAATAGATGTCAATCGTATGCTATCAACTTTCAAACCCCAGCAGAGAAAATGTTTGGAAACAAAGACTTAACAAGACTTAGGATATTTGGATCAAAATCATGGGTTCATATCATACCAAAACAAGATAAACTTTTACAAAGAGCTAAAGAAATGAGACTCGTAGGATATAGCCCAAATGGATATCGTCTATGGGACCCTACAAGTAATAAAGTGATAGTGTCAAGAGATGTCAGAATTGATGAAACGCAATTAAACTACAAGGAAACTGAAGAAAAGCATAACATAAAAGGAGAAACGTACTATGAAGATGAATTAATACAAAATGACGATACTGAGAAGGACACAGAAGCTGACCAAGAGACAAGAATACCTAATGAGAATGATGAAAACATGCAACATGATAGAATACCTGATAAAACAAATAAAGAAGTATCAAAGAATAATCAAGATGATGAAGAAGAAAATGTAAAATTGAATAAAAAAGGTAGAATAAATATACAGAAATATATAACAAGATCAGGAAGAGAAATTCGCAAACCAAAAAACTTAGAAAAATATGATCTAAATATAGCATACTGTCTTTTATCGGGAGACCCACAAGAATTTGAAGATGCAATTCATAACAAAGACTGGGAAAAAGCAATAAAGACAATTAAATTCACAAGTGAAATTAGAGACATGGGAAGAAGCTACATTACCAATGGGAAGTAA
Protein
MHSDTEQDFYAVSRFPRVLGTLDGTHIRIQSPCSQIEEEFRNRKGYFSLNVQAVCSNADLLFMNVVARWPGSAHDATIFNNSDLRAQCENGSFGNKWLLGDSAYPLKPYLLTPLINTQTRGQQLYNEAHIRTRNCIERCFGVWKRRFPVVALTLRLSLLRANAVIIATAVLHNICRLKKLDDISPEVEIPEADVLPTESSTEETFKEKECKEAIEDGITEEILKSEKFKKKDAKARSLIVNCLSDKHLEYVRSATSAKEMIENLRKIFKRKSTLSALYVRKKLLTLKCDPSVELSDHFNNFDMLIRQLEETGSTVNEQDKVCHLLLTMPDSYNTTIAALETTNVELTTEYVKSKLLDAELKNQNNSIESQDAHSFLSCFKCGKTGHKAIQCRTGHLMRGQHRGKMGNVGYFNGSNTRDFYRGNGRDFNPGNGRGFNRGNPRGNVRGQSRGSSRDIIPQQGKSALSERNPFSFVASEKVLSVDVYNEDIKFIIDSGASQNLVIDSYEKYMSNIEYLETKTKIYVANGQYLFSSKKGILNVKYNNLPIKIEALLVKGLSHNLLSVKKLLEKGNSVRFHKNSVSIAKGNNIIYGTMLNSLYAINLTLNLEKCYSINNDKDLWHKRLGHANRKHLKLLKLPISEKPCGICVEGKSTRLPFSTTPRPRSKYIGELIHTDISGPINIPTLTNEVYFHTIIDDYTHFCEVYLLQRKSEATDRLIEYVNRMERQIECKVKKIRSDNGGEFKNEKLNKFCKDKGIVQQFTLPYSPQSNGVSERMNRNIYNRARTLLIESGLPKTLWGEAVRCAVYQTNRCQSYAINFQTPAEKMFGNKDLTRLRIFGSKSWVHIIPKQDKLLQRAKEMRLVGYSPNGYRLWDPTSNKVIVSRDVRIDETQLNYKETEEKHNIKGETYYEDELIQNDDTEKDTEADQETRIPNENDENMQHDRIPDKTNKEVSKNNQDDEEENVKLNKKGRINIQKYITRSGREIRKPKNLEKYDLNIAYCLLSGDPQEFEDAIHNKDWEKAIKTIKFTSEIRDMGRSYITNGK
Summary
Uniprot
Pubmed
EMBL
AB014676
BAA74713.1
GBHO01027564
JAG16040.1
GDHC01015802
JAQ02827.1
+ More
GBHO01034936 JAG08668.1 GBHO01002616 JAG40988.1 GBHO01035173 JAG08431.1 GEDC01012841 GEDC01000503 JAS24457.1 JAS36795.1 LBMM01007046 KMQ90088.1 GEZM01072005 JAV65635.1 GEZM01028916 JAV86195.1 JXUM01085595 KQ563513 KXJ73717.1 JAV86196.1 ABLF02041793 GAKP01015342 JAC43610.1 LBMM01010924 KMQ87150.1 GBBI01004033 JAC14679.1 GDHC01007678 JAQ10951.1 GDUN01000694 JAN95225.1 LBMM01012772 KMQ86003.1 GECL01003258 JAP02866.1 GEHC01000879 JAV46766.1 GDHF01030254 JAI22060.1 D83003 BAA11674.1 GBXI01002679 JAD11613.1 BABH01010608
GBHO01034936 JAG08668.1 GBHO01002616 JAG40988.1 GBHO01035173 JAG08431.1 GEDC01012841 GEDC01000503 JAS24457.1 JAS36795.1 LBMM01007046 KMQ90088.1 GEZM01072005 JAV65635.1 GEZM01028916 JAV86195.1 JXUM01085595 KQ563513 KXJ73717.1 JAV86196.1 ABLF02041793 GAKP01015342 JAC43610.1 LBMM01010924 KMQ87150.1 GBBI01004033 JAC14679.1 GDHC01007678 JAQ10951.1 GDUN01000694 JAN95225.1 LBMM01012772 KMQ86003.1 GECL01003258 JAP02866.1 GEHC01000879 JAV46766.1 GDHF01030254 JAI22060.1 D83003 BAA11674.1 GBXI01002679 JAD11613.1 BABH01010608
Proteomes
Interpro
Gene 3D
ProteinModelPortal
PDB
5UOQ
E-value=4.92891e-08,
Score=141
Ontologies
GO
PANTHER
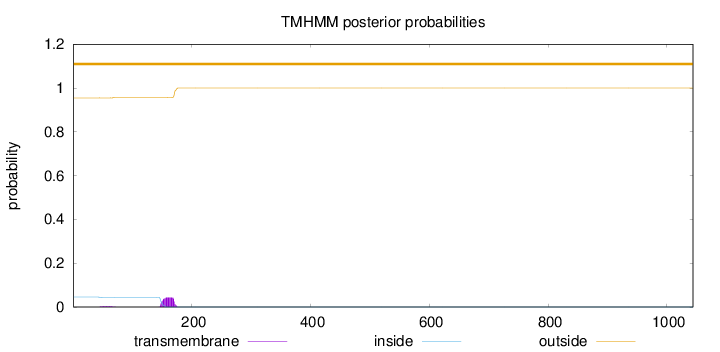
Topology
Length:
1044
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.00239
Exp number, first 60 AAs:
0.03724
Total prob of N-in:
0.04510
outside
1 - 1044
Population Genetic Test Statistics
Pi
297.791392
Theta
175.864807
Tajima's D
2.044731
CLR
12.270604
CSRT
0.886555672216389
Interpretation
Uncertain
