Gene
KWMTBOMO12133
Pre Gene Modal
BGIBMGA005185
Annotation
copia_protein_[Lasius_niger]
Location in the cell
Nuclear Reliability : 3.614
Sequence
CDS
ATGGAAGTAAATAATACCACTCATGTGATGAAACTGGAAGGTGCGAAAAATTGGAATGTTTGGAAGTTTCAAATAACAGTATTGTTGAAAGGTCAAGGATGGTTGGACGTTGTTGAAGGGCGGACGGTAAAACCAGAAGACGCTGTTGCAAGAGCTGCCTGGGAAGTAAAGGATGCTAAGGCTCAATCATTGATAGTCACCAGGATGTCTGAGGATGTCATGCTGCATATCATAACTTGTACATCTTCTGCTGAAATGTGGCAAAAGCTGTTATGTGTCTACGAACAAAAGTCGGAAACCAGCATTCACATTATACAGCAACGTTTCTTTCAATATAAATATGAGAAAGGAGTTGATATGTCGGTGTTTATATCTAAAATCCAAGAAATGCAGAATGAATATAAACATTTTATATCGGCATGGGAGTCCGCACCCGATAATAAACAAACATTTGATAATTTGGTGGCAAGGCTATTAGTAGAAGAGGAAAGGTTGAAGGAAAAGATTGAAGAAAACCAGGAGACGACGTCAGCGTTCGTAGCCAAGAGAAATGCTAAGTGTTTTAAATGTGGCAAACCGGGACATTACCAGTCAGAGTGTCGAAATAATAGAGACAATGAGAAAAACGTGGACTTTAATAAGTGCTTTTATTGTGGTAAACCGGGACATGTTAAGGCGCAGTGTCGTTATAGAAAGCTAAAGGACAATAAGAAAATGAGTAATGCGTTCGTTGTGTGTAACTCAGAAGACGAAAAATATGAAAAATCAAAGTGGCTAGTTGATTCAGGGGCTAGCGAACATATGTGCTTTGATAGTGAGCATGTAAATGTAGCTAAAGTAGTAAAGAATGACTTATATAAATGGCATCAGAAGTTAGCACATCAGAACTTTGATCACGTAAAGAAAGTGTTGGCGAAAAACAACATTGTTGTGAAACAAACCAATGTGCCACAATGTGAAAGCTGCTTAAAAGGAAAAATACATAGGCTACCATTTATGAAAAGCGATAGTGTAAGCACTAGGACATGCGAATTAATACATGGAGACACATGTGGACCCATGGAAGAAGAGTCCATTGGTGGTGAGAACACATATTTCAAAAAAAAAAGGCAGAATGCTGATTTGGAAAGAAAAAAGCTGATGATTGAGATATTAAACCTCAAAAAAGAGCATGAACTTCGGATGCAGATTCTAAAAGCAAAACAGTGGAGACTAGAACAACATTAG
Protein
MEVNNTTHVMKLEGAKNWNVWKFQITVLLKGQGWLDVVEGRTVKPEDAVARAAWEVKDAKAQSLIVTRMSEDVMLHIITCTSSAEMWQKLLCVYEQKSETSIHIIQQRFFQYKYEKGVDMSVFISKIQEMQNEYKHFISAWESAPDNKQTFDNLVARLLVEEERLKEKIEENQETTSAFVAKRNAKCFKCGKPGHYQSECRNNRDNEKNVDFNKCFYCGKPGHVKAQCRYRKLKDNKKMSNAFVVCNSEDEKYEKSKWLVDSGASEHMCFDSEHVNVAKVVKNDLYKWHQKLAHQNFDHVKKVLAKNNIVVKQTNVPQCESCLKGKIHRLPFMKSDSVSTRTCELIHGDTCGPMEEESIGGENTYFKKKRQNADLERKKLMIEILNLKKEHELRMQILKAKQWRLEQH
Summary
Uniprot
A0A2A4JP38
A0A2A4JBE6
A0A1Y1LCN3
A0A2R7X964
A0A2R7W1Y3
A0A1Y1K3Y8
+ More
A0A1Y1K1A3 X1XSG1 A0A1Y1NEM6 A0A2S2P896 J9LCG1 A0A151II00 X1XN05 X1WXE3 A0A1Y1L0K1 X1WMK8 A0A1Y1KZM0 A0A2J7QWT7 A0A0J7K689 V5FYN4 X1X3B7 X1WT66 X1WZ36 A0A0J7K717 A0A151J9U8 X1WMG8 A0A3L8DP62 X1XQX4 A0A0P6CUX5 A0A0A9XEU3 A0A1Y1N4J3 A0A226DW10 A0A226DUK4 A0A0J7N723 A0A151IWP6 A0A1Y1N6B6 A0A0J7KDE2 A0A0P6CCJ8 A0A195F4T3 A0A0A9XY62 K7J7F1 A0A195FFU8 A0A0A9Y466 K7JGG9 K7JAW3 X1X2T1 A0A0J7KAF6 K7J987 A0A0J7N8C0 A0A1B6FK59 A0A1Y1K2V0 A0A151ITN2 A0A0J7NDW8 A0A085LL10 A0A1Y1LRT7 A0A0V0ULL1
A0A1Y1K1A3 X1XSG1 A0A1Y1NEM6 A0A2S2P896 J9LCG1 A0A151II00 X1XN05 X1WXE3 A0A1Y1L0K1 X1WMK8 A0A1Y1KZM0 A0A2J7QWT7 A0A0J7K689 V5FYN4 X1X3B7 X1WT66 X1WZ36 A0A0J7K717 A0A151J9U8 X1WMG8 A0A3L8DP62 X1XQX4 A0A0P6CUX5 A0A0A9XEU3 A0A1Y1N4J3 A0A226DW10 A0A226DUK4 A0A0J7N723 A0A151IWP6 A0A1Y1N6B6 A0A0J7KDE2 A0A0P6CCJ8 A0A195F4T3 A0A0A9XY62 K7J7F1 A0A195FFU8 A0A0A9Y466 K7JGG9 K7JAW3 X1X2T1 A0A0J7KAF6 K7J987 A0A0J7N8C0 A0A1B6FK59 A0A1Y1K2V0 A0A151ITN2 A0A0J7NDW8 A0A085LL10 A0A1Y1LRT7 A0A0V0ULL1
EMBL
NWSH01000988
PCG73213.1
NWSH01002023
PCG69395.1
GEZM01063091
JAV69316.1
+ More
KK863047 PTY28326.1 KK854253 PTY13757.1 GEZM01096003 JAV55208.1 GEZM01096000 JAV55213.1 ABLF02042028 GEZM01005616 JAV96038.1 GGMR01012789 MBY25408.1 ABLF02020242 ABLF02020246 ABLF02042026 KQ977592 KYN01654.1 ABLF02041733 ABLF02060404 ABLF02007537 ABLF02066133 GEZM01073374 JAV65136.1 ABLF02066512 JAV65135.1 NEVH01009420 PNF33053.1 LBMM01012772 KMQ86003.1 GALX01005676 JAB62790.1 ABLF02042030 ABLF02016448 ABLF02056079 ABLF02056084 LBMM01012696 KMQ86054.1 KQ979396 KYN21742.1 ABLF02057951 QOIP01000006 RLU22076.1 GDIP01002833 JAN00883.1 GBHO01024362 JAG19242.1 GEZM01016434 JAV91276.1 LNIX01000011 OXA48994.1 OXA48899.1 LBMM01008978 KMQ88490.1 KQ980862 KYN12127.1 GEZM01013586 GEZM01013585 JAV92450.1 LBMM01008998 KMQ88473.1 GDIP01002579 JAN01137.1 KQ981826 KYN35089.1 GBHO01019278 JAG24326.1 AAZX01012276 KQ981617 KYN39263.1 GBHO01019284 JAG24320.1 AAZX01000214 AAZX01007161 AAZX01023229 ABLF02003699 ABLF02003700 ABLF02007662 ABLF02007663 ABLF02019401 ABLF02049974 ABLF02050034 LBMM01010759 KMQ87259.1 AAZX01006867 LBMM01008433 KMQ88885.1 GECZ01019212 JAS50557.1 GEZM01098859 JAV53706.1 KQ981003 KYN10396.1 LBMM01006309 KMQ90715.1 KL363445 KFD45656.1 GEZM01048830 JAV76369.1 JYDK01000940 JYDK01000790 KRX51539.1 KRX53067.1
KK863047 PTY28326.1 KK854253 PTY13757.1 GEZM01096003 JAV55208.1 GEZM01096000 JAV55213.1 ABLF02042028 GEZM01005616 JAV96038.1 GGMR01012789 MBY25408.1 ABLF02020242 ABLF02020246 ABLF02042026 KQ977592 KYN01654.1 ABLF02041733 ABLF02060404 ABLF02007537 ABLF02066133 GEZM01073374 JAV65136.1 ABLF02066512 JAV65135.1 NEVH01009420 PNF33053.1 LBMM01012772 KMQ86003.1 GALX01005676 JAB62790.1 ABLF02042030 ABLF02016448 ABLF02056079 ABLF02056084 LBMM01012696 KMQ86054.1 KQ979396 KYN21742.1 ABLF02057951 QOIP01000006 RLU22076.1 GDIP01002833 JAN00883.1 GBHO01024362 JAG19242.1 GEZM01016434 JAV91276.1 LNIX01000011 OXA48994.1 OXA48899.1 LBMM01008978 KMQ88490.1 KQ980862 KYN12127.1 GEZM01013586 GEZM01013585 JAV92450.1 LBMM01008998 KMQ88473.1 GDIP01002579 JAN01137.1 KQ981826 KYN35089.1 GBHO01019278 JAG24326.1 AAZX01012276 KQ981617 KYN39263.1 GBHO01019284 JAG24320.1 AAZX01000214 AAZX01007161 AAZX01023229 ABLF02003699 ABLF02003700 ABLF02007662 ABLF02007663 ABLF02019401 ABLF02049974 ABLF02050034 LBMM01010759 KMQ87259.1 AAZX01006867 LBMM01008433 KMQ88885.1 GECZ01019212 JAS50557.1 GEZM01098859 JAV53706.1 KQ981003 KYN10396.1 LBMM01006309 KMQ90715.1 KL363445 KFD45656.1 GEZM01048830 JAV76369.1 JYDK01000940 JYDK01000790 KRX51539.1 KRX53067.1
Proteomes
Pfam
Interpro
Gene 3D
ProteinModelPortal
A0A2A4JP38
A0A2A4JBE6
A0A1Y1LCN3
A0A2R7X964
A0A2R7W1Y3
A0A1Y1K3Y8
+ More
A0A1Y1K1A3 X1XSG1 A0A1Y1NEM6 A0A2S2P896 J9LCG1 A0A151II00 X1XN05 X1WXE3 A0A1Y1L0K1 X1WMK8 A0A1Y1KZM0 A0A2J7QWT7 A0A0J7K689 V5FYN4 X1X3B7 X1WT66 X1WZ36 A0A0J7K717 A0A151J9U8 X1WMG8 A0A3L8DP62 X1XQX4 A0A0P6CUX5 A0A0A9XEU3 A0A1Y1N4J3 A0A226DW10 A0A226DUK4 A0A0J7N723 A0A151IWP6 A0A1Y1N6B6 A0A0J7KDE2 A0A0P6CCJ8 A0A195F4T3 A0A0A9XY62 K7J7F1 A0A195FFU8 A0A0A9Y466 K7JGG9 K7JAW3 X1X2T1 A0A0J7KAF6 K7J987 A0A0J7N8C0 A0A1B6FK59 A0A1Y1K2V0 A0A151ITN2 A0A0J7NDW8 A0A085LL10 A0A1Y1LRT7 A0A0V0ULL1
A0A1Y1K1A3 X1XSG1 A0A1Y1NEM6 A0A2S2P896 J9LCG1 A0A151II00 X1XN05 X1WXE3 A0A1Y1L0K1 X1WMK8 A0A1Y1KZM0 A0A2J7QWT7 A0A0J7K689 V5FYN4 X1X3B7 X1WT66 X1WZ36 A0A0J7K717 A0A151J9U8 X1WMG8 A0A3L8DP62 X1XQX4 A0A0P6CUX5 A0A0A9XEU3 A0A1Y1N4J3 A0A226DW10 A0A226DUK4 A0A0J7N723 A0A151IWP6 A0A1Y1N6B6 A0A0J7KDE2 A0A0P6CCJ8 A0A195F4T3 A0A0A9XY62 K7J7F1 A0A195FFU8 A0A0A9Y466 K7JGG9 K7JAW3 X1X2T1 A0A0J7KAF6 K7J987 A0A0J7N8C0 A0A1B6FK59 A0A1Y1K2V0 A0A151ITN2 A0A0J7NDW8 A0A085LL10 A0A1Y1LRT7 A0A0V0ULL1
PDB
5A9E
E-value=0.000108852,
Score=108
Ontologies
GO
PANTHER
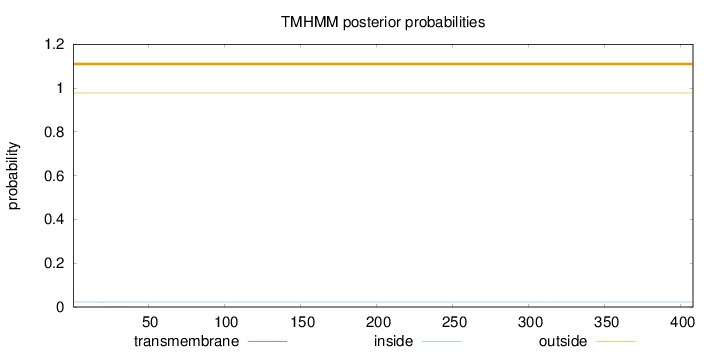
Topology
Length:
408
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00438
Exp number, first 60 AAs:
0.00356
Total prob of N-in:
0.02264
outside
1 - 408
Population Genetic Test Statistics
Pi
137.219215
Theta
158.444921
Tajima's D
-0.418835
CLR
0.594621
CSRT
0.255887205639718
Interpretation
Uncertain
