Pre Gene Modal
BGIBMGA004241
Annotation
putative_importin_[Danaus_plexippus]
Location in the cell
Cytoplasmic Reliability : 1.69
Sequence
CDS
ATGGATGTAACCGAAGAAAATTTGGCAACTCTAGCAAACTACTTACAGCAAACATTGAATCCAGATCAGAACATCAGAAGACCCGCTGAGAAATTTTTAGAGAGTGTAGAAGTCAACCAGAACTATGCCATACTATTGTTGCATCTCATTGACAAGAACAACTATGATAATACAATTAGAGTGGCAGCAGCAATTGTGTTTAAAAACTATATTAAACGGAATTGGCCTATTGAAGAAGACGGCGTGGATCGCATTCATATGGCAGACAGAGTCTCTATCAAGAATCTCATAGTGAACTTGATGCTTAAATCACCAGAAGCAATACAGAAACAATTTAGTGATGCCGTTTCCATAATCGGAAAATATGATTTTCCTGAGAAGTGGCCAACACTTATACCAGAAATGGTGGAAAAATTTGCTACAGGCGACTTCCATGTGATTAATGGTATACTACGGACAGCTCATTCATTATTCAAGAGGTACAGATACGAATTTAAATCTCAAAAACTATGGGAGGAAATCAAACATGTCCTTGAGAATTTTGCAAGACCCCTCACAGATCTCTTTGTTGCTACGATAGAACTAACTCAGAAGCATGCATCGGACGTCCAGGCCCTGCGTGTAATCTATGGGTCCCTCGTCCTCATCTGCAAAGTGTTCTATTCACTTAACTACCAGGATCTGCCGGAGTTCTTTGAAGACAACATGCCCATTTGGATGCCAAATCTGCTGAGACTACTGCAAGTGGATGTTCCGAATGTTGATGATGGAGACGACGACAACCCTGGACTAATGGAGAGACTCCGCACGGAGATATGCGAATGCGTTTCGTTATACGCACTGAAGTACGAGGAGGAGTTTTTGCCTTATTCAAGCGGCTTCGTGACGGCTGTGTGGAACATACTCCTCGCCACAACGCAGAAAGCTAAATACGATATGGTAACTATGTATTATTAA
Protein
MDVTEENLATLANYLQQTLNPDQNIRRPAEKFLESVEVNQNYAILLLHLIDKNNYDNTIRVAAAIVFKNYIKRNWPIEEDGVDRIHMADRVSIKNLIVNLMLKSPEAIQKQFSDAVSIIGKYDFPEKWPTLIPEMVEKFATGDFHVINGILRTAHSLFKRYRYEFKSQKLWEEIKHVLENFARPLTDLFVATIELTQKHASDVQALRVIYGSLVLICKVFYSLNYQDLPEFFEDNMPIWMPNLLRLLQVDVPNVDDGDDDNPGLMERLRTEICECVSLYALKYEEEFLPYSSGFVTAVWNILLATTQKAKYDMVTMYY
Summary
Uniprot
H9J402
A0A2A4KB88
A0A212F558
A0A194QFC7
A0A2W1BFF6
A0A2P8XM62
+ More
E2A3P1 E9IFD4 A0A151IJS9 A0A067REG1 A0A158NKK0 A0A195B0R1 A0A195FBK4 E2BZ56 A0A151JBE9 A0A151WRQ7 A0A3L8D5Z4 A0A1Y1KAV5 D6WTV7 F4WWC9 A0A1Z5KV73 U4U9R7 N6U7H0 A0A2A3E9T1 A0A088A291 K7IQA9 L7MEW5 A0A023GFX0 A0A131Y3V5 A0A2D1QUE9 A0A147BS71 A0A2J7RI15 A0A232EYL1 A0A1B0CYT2 A0A2J7RI16 A0A1L8DKS7 A0A2R5LMM5 A0A224YY17 A0A1B0CRX9 A0A131Z471 A0A1B6CNE8 A0A310SPL4 A0A131XJZ1 A0A0A1XDD7 A0A026WED2 T1PBI4 A0A0C9R0E6 T1HSJ8 A0A0L0BSZ8 A0A1I8NSS0 A0A034W6I8 A0A1S3IIH3 A0A182J0K5 A0A182QT46 A0A0K8V1J2 A0A293N7H1 A0A0P5JKN8 A0A182Y9N4 A0A0P5UIL6 A0A182V1L9 A0A0P5QAX0 A0A1E1XAG0 A0A182M1Y2 A0A084WNJ8 A0A182U6G5 A0A182WXX2 Q7QE09 A0A182I5Z3 A0A182KV25 A0A182PM17 A0A0P5PEE1 A0A182K3X1 A0A069DXB6 A0A2H1VPK9 A0A1B0A4R0 A0A1B0G9Q1 A0A1Q3F0Q1 A0A182WE55 A0A182RW86 A0A0P5YUF4 A0A1A9XVX2 A0A1A9VIB6 A0A023FA40 A0A224XHZ2 A0A182H483 A0A1B6GBG3 A0A0P4VSH5 W8BRV6 A0A210Q9G8 E9HY45 A0A1W4VY12 B4MYY0 A0A0P4Y7H2 B4KJR4 B4JQX2 A0A0P5ZVT1 B3NNL6 A0A0P5U809 B4I558 B4Q7H1
E2A3P1 E9IFD4 A0A151IJS9 A0A067REG1 A0A158NKK0 A0A195B0R1 A0A195FBK4 E2BZ56 A0A151JBE9 A0A151WRQ7 A0A3L8D5Z4 A0A1Y1KAV5 D6WTV7 F4WWC9 A0A1Z5KV73 U4U9R7 N6U7H0 A0A2A3E9T1 A0A088A291 K7IQA9 L7MEW5 A0A023GFX0 A0A131Y3V5 A0A2D1QUE9 A0A147BS71 A0A2J7RI15 A0A232EYL1 A0A1B0CYT2 A0A2J7RI16 A0A1L8DKS7 A0A2R5LMM5 A0A224YY17 A0A1B0CRX9 A0A131Z471 A0A1B6CNE8 A0A310SPL4 A0A131XJZ1 A0A0A1XDD7 A0A026WED2 T1PBI4 A0A0C9R0E6 T1HSJ8 A0A0L0BSZ8 A0A1I8NSS0 A0A034W6I8 A0A1S3IIH3 A0A182J0K5 A0A182QT46 A0A0K8V1J2 A0A293N7H1 A0A0P5JKN8 A0A182Y9N4 A0A0P5UIL6 A0A182V1L9 A0A0P5QAX0 A0A1E1XAG0 A0A182M1Y2 A0A084WNJ8 A0A182U6G5 A0A182WXX2 Q7QE09 A0A182I5Z3 A0A182KV25 A0A182PM17 A0A0P5PEE1 A0A182K3X1 A0A069DXB6 A0A2H1VPK9 A0A1B0A4R0 A0A1B0G9Q1 A0A1Q3F0Q1 A0A182WE55 A0A182RW86 A0A0P5YUF4 A0A1A9XVX2 A0A1A9VIB6 A0A023FA40 A0A224XHZ2 A0A182H483 A0A1B6GBG3 A0A0P4VSH5 W8BRV6 A0A210Q9G8 E9HY45 A0A1W4VY12 B4MYY0 A0A0P4Y7H2 B4KJR4 B4JQX2 A0A0P5ZVT1 B3NNL6 A0A0P5U809 B4I558 B4Q7H1
Pubmed
19121390
22118469
26354079
28756777
29403074
20798317
+ More
21282665 24845553 21347285 30249741 28004739 18362917 19820115 21719571 28528879 23537049 20075255 25576852 29652888 28648823 28797301 26830274 28049606 25830018 24508170 25315136 26108605 25348373 25244985 28503490 24438588 12364791 14747013 17210077 20966253 26334808 25474469 26483478 27129103 24495485 28812685 21292972 17994087 22936249
21282665 24845553 21347285 30249741 28004739 18362917 19820115 21719571 28528879 23537049 20075255 25576852 29652888 28648823 28797301 26830274 28049606 25830018 24508170 25315136 26108605 25348373 25244985 28503490 24438588 12364791 14747013 17210077 20966253 26334808 25474469 26483478 27129103 24495485 28812685 21292972 17994087 22936249
EMBL
BABH01040156
BABH01040157
BABH01040158
BABH01040159
BABH01040160
NWSH01000004
+ More
PCG81010.1 AGBW02010240 OWR48872.1 KQ459053 KPJ04187.1 KZ150239 PZC71937.1 PYGN01001737 PSN33093.1 GL436457 EFN71933.1 GL762838 EFZ20732.1 KQ977294 KYN03892.1 KK852686 KDR18464.1 ADTU01018869 KQ976692 KYM77882.1 KQ981693 KYN37793.1 GL451563 EFN79025.1 KQ979182 KYN22319.1 KQ982805 KYQ50487.1 QOIP01000013 RLU15268.1 GEZM01087742 GEZM01087741 JAV58514.1 KQ971352 EFA07339.1 GL888406 EGI61471.1 GFJQ02007981 JAV98988.1 KB632188 ERL89802.1 APGK01045816 KB741039 ENN74537.1 KZ288311 PBC28487.1 GACK01003231 JAA61803.1 GBBM01003540 JAC31878.1 GEFM01002746 JAP73050.1 KY285054 ATP16157.1 GEGO01001795 JAR93609.1 NEVH01003506 PNF40477.1 NNAY01001630 OXU23377.1 AJVK01009334 AJVK01009335 AJVK01009336 PNF40478.1 GFDF01007018 JAV07066.1 GGLE01006654 MBY10780.1 GFPF01007718 MAA18864.1 AJWK01025448 GEDV01002358 JAP86199.1 GEDC01022433 JAS14865.1 KQ761976 OAD56402.1 GEFH01001899 JAP66682.1 GBXI01007135 GBXI01005322 JAD07157.1 JAD08970.1 KK107249 EZA54460.1 KA646122 AFP60751.1 GBYB01001470 JAG71237.1 ACPB03005913 ACPB03005914 JRES01001418 KNC23108.1 GAKP01009554 JAC49398.1 AXCN02001080 GDHF01019545 JAI32769.1 GFWV01022843 MAA47570.1 GDIQ01201645 JAK50080.1 GDIP01113827 JAL89887.1 GDIQ01138366 JAL13360.1 GFAC01002943 JAT96245.1 AXCM01007239 ATLV01024618 KE525353 KFB51792.1 AAAB01008848 EAA07074.1 APCN01003537 GDIQ01135820 JAL15906.1 GBGD01000349 JAC88540.1 ODYU01003688 SOQ42763.1 CCAG010003515 GFDL01013936 JAV21109.1 GDIP01066578 JAM37137.1 GBBI01000365 JAC18347.1 GFTR01008356 JAW08070.1 JXUM01108986 KQ565280 KXJ71133.1 GECZ01010007 JAS59762.1 GDKW01001181 JAI55414.1 GAMC01010569 JAB95986.1 NEDP02004514 OWF45371.1 GL733109 EFX63333.1 CH963913 EDW77319.1 GDIP01233710 JAI89691.1 CH933807 EDW11509.1 CH916372 EDV99302.1 GDIP01038690 JAM65025.1 CH954179 EDV56667.1 GDIP01118334 JAL85380.1 CH480822 EDW55514.1 CM000361 CM002910 EDX05269.1 KMY90614.1
PCG81010.1 AGBW02010240 OWR48872.1 KQ459053 KPJ04187.1 KZ150239 PZC71937.1 PYGN01001737 PSN33093.1 GL436457 EFN71933.1 GL762838 EFZ20732.1 KQ977294 KYN03892.1 KK852686 KDR18464.1 ADTU01018869 KQ976692 KYM77882.1 KQ981693 KYN37793.1 GL451563 EFN79025.1 KQ979182 KYN22319.1 KQ982805 KYQ50487.1 QOIP01000013 RLU15268.1 GEZM01087742 GEZM01087741 JAV58514.1 KQ971352 EFA07339.1 GL888406 EGI61471.1 GFJQ02007981 JAV98988.1 KB632188 ERL89802.1 APGK01045816 KB741039 ENN74537.1 KZ288311 PBC28487.1 GACK01003231 JAA61803.1 GBBM01003540 JAC31878.1 GEFM01002746 JAP73050.1 KY285054 ATP16157.1 GEGO01001795 JAR93609.1 NEVH01003506 PNF40477.1 NNAY01001630 OXU23377.1 AJVK01009334 AJVK01009335 AJVK01009336 PNF40478.1 GFDF01007018 JAV07066.1 GGLE01006654 MBY10780.1 GFPF01007718 MAA18864.1 AJWK01025448 GEDV01002358 JAP86199.1 GEDC01022433 JAS14865.1 KQ761976 OAD56402.1 GEFH01001899 JAP66682.1 GBXI01007135 GBXI01005322 JAD07157.1 JAD08970.1 KK107249 EZA54460.1 KA646122 AFP60751.1 GBYB01001470 JAG71237.1 ACPB03005913 ACPB03005914 JRES01001418 KNC23108.1 GAKP01009554 JAC49398.1 AXCN02001080 GDHF01019545 JAI32769.1 GFWV01022843 MAA47570.1 GDIQ01201645 JAK50080.1 GDIP01113827 JAL89887.1 GDIQ01138366 JAL13360.1 GFAC01002943 JAT96245.1 AXCM01007239 ATLV01024618 KE525353 KFB51792.1 AAAB01008848 EAA07074.1 APCN01003537 GDIQ01135820 JAL15906.1 GBGD01000349 JAC88540.1 ODYU01003688 SOQ42763.1 CCAG010003515 GFDL01013936 JAV21109.1 GDIP01066578 JAM37137.1 GBBI01000365 JAC18347.1 GFTR01008356 JAW08070.1 JXUM01108986 KQ565280 KXJ71133.1 GECZ01010007 JAS59762.1 GDKW01001181 JAI55414.1 GAMC01010569 JAB95986.1 NEDP02004514 OWF45371.1 GL733109 EFX63333.1 CH963913 EDW77319.1 GDIP01233710 JAI89691.1 CH933807 EDW11509.1 CH916372 EDV99302.1 GDIP01038690 JAM65025.1 CH954179 EDV56667.1 GDIP01118334 JAL85380.1 CH480822 EDW55514.1 CM000361 CM002910 EDX05269.1 KMY90614.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053268
UP000245037
UP000000311
+ More
UP000078542 UP000027135 UP000005205 UP000078540 UP000078541 UP000008237 UP000078492 UP000075809 UP000279307 UP000007266 UP000007755 UP000030742 UP000019118 UP000242457 UP000005203 UP000002358 UP000235965 UP000215335 UP000092462 UP000092461 UP000053097 UP000095301 UP000015103 UP000037069 UP000095300 UP000085678 UP000075880 UP000075886 UP000076408 UP000075903 UP000075883 UP000030765 UP000075902 UP000076407 UP000007062 UP000075840 UP000075882 UP000075885 UP000075881 UP000092445 UP000092444 UP000075920 UP000075900 UP000092443 UP000078200 UP000069940 UP000249989 UP000242188 UP000000305 UP000192221 UP000007798 UP000009192 UP000001070 UP000008711 UP000001292 UP000000304
UP000078542 UP000027135 UP000005205 UP000078540 UP000078541 UP000008237 UP000078492 UP000075809 UP000279307 UP000007266 UP000007755 UP000030742 UP000019118 UP000242457 UP000005203 UP000002358 UP000235965 UP000215335 UP000092462 UP000092461 UP000053097 UP000095301 UP000015103 UP000037069 UP000095300 UP000085678 UP000075880 UP000075886 UP000076408 UP000075903 UP000075883 UP000030765 UP000075902 UP000076407 UP000007062 UP000075840 UP000075882 UP000075885 UP000075881 UP000092445 UP000092444 UP000075920 UP000075900 UP000092443 UP000078200 UP000069940 UP000249989 UP000242188 UP000000305 UP000192221 UP000007798 UP000009192 UP000001070 UP000008711 UP000001292 UP000000304
PRIDE
Interpro
SUPFAM
SSF48371
SSF48371
Gene 3D
ProteinModelPortal
H9J402
A0A2A4KB88
A0A212F558
A0A194QFC7
A0A2W1BFF6
A0A2P8XM62
+ More
E2A3P1 E9IFD4 A0A151IJS9 A0A067REG1 A0A158NKK0 A0A195B0R1 A0A195FBK4 E2BZ56 A0A151JBE9 A0A151WRQ7 A0A3L8D5Z4 A0A1Y1KAV5 D6WTV7 F4WWC9 A0A1Z5KV73 U4U9R7 N6U7H0 A0A2A3E9T1 A0A088A291 K7IQA9 L7MEW5 A0A023GFX0 A0A131Y3V5 A0A2D1QUE9 A0A147BS71 A0A2J7RI15 A0A232EYL1 A0A1B0CYT2 A0A2J7RI16 A0A1L8DKS7 A0A2R5LMM5 A0A224YY17 A0A1B0CRX9 A0A131Z471 A0A1B6CNE8 A0A310SPL4 A0A131XJZ1 A0A0A1XDD7 A0A026WED2 T1PBI4 A0A0C9R0E6 T1HSJ8 A0A0L0BSZ8 A0A1I8NSS0 A0A034W6I8 A0A1S3IIH3 A0A182J0K5 A0A182QT46 A0A0K8V1J2 A0A293N7H1 A0A0P5JKN8 A0A182Y9N4 A0A0P5UIL6 A0A182V1L9 A0A0P5QAX0 A0A1E1XAG0 A0A182M1Y2 A0A084WNJ8 A0A182U6G5 A0A182WXX2 Q7QE09 A0A182I5Z3 A0A182KV25 A0A182PM17 A0A0P5PEE1 A0A182K3X1 A0A069DXB6 A0A2H1VPK9 A0A1B0A4R0 A0A1B0G9Q1 A0A1Q3F0Q1 A0A182WE55 A0A182RW86 A0A0P5YUF4 A0A1A9XVX2 A0A1A9VIB6 A0A023FA40 A0A224XHZ2 A0A182H483 A0A1B6GBG3 A0A0P4VSH5 W8BRV6 A0A210Q9G8 E9HY45 A0A1W4VY12 B4MYY0 A0A0P4Y7H2 B4KJR4 B4JQX2 A0A0P5ZVT1 B3NNL6 A0A0P5U809 B4I558 B4Q7H1
E2A3P1 E9IFD4 A0A151IJS9 A0A067REG1 A0A158NKK0 A0A195B0R1 A0A195FBK4 E2BZ56 A0A151JBE9 A0A151WRQ7 A0A3L8D5Z4 A0A1Y1KAV5 D6WTV7 F4WWC9 A0A1Z5KV73 U4U9R7 N6U7H0 A0A2A3E9T1 A0A088A291 K7IQA9 L7MEW5 A0A023GFX0 A0A131Y3V5 A0A2D1QUE9 A0A147BS71 A0A2J7RI15 A0A232EYL1 A0A1B0CYT2 A0A2J7RI16 A0A1L8DKS7 A0A2R5LMM5 A0A224YY17 A0A1B0CRX9 A0A131Z471 A0A1B6CNE8 A0A310SPL4 A0A131XJZ1 A0A0A1XDD7 A0A026WED2 T1PBI4 A0A0C9R0E6 T1HSJ8 A0A0L0BSZ8 A0A1I8NSS0 A0A034W6I8 A0A1S3IIH3 A0A182J0K5 A0A182QT46 A0A0K8V1J2 A0A293N7H1 A0A0P5JKN8 A0A182Y9N4 A0A0P5UIL6 A0A182V1L9 A0A0P5QAX0 A0A1E1XAG0 A0A182M1Y2 A0A084WNJ8 A0A182U6G5 A0A182WXX2 Q7QE09 A0A182I5Z3 A0A182KV25 A0A182PM17 A0A0P5PEE1 A0A182K3X1 A0A069DXB6 A0A2H1VPK9 A0A1B0A4R0 A0A1B0G9Q1 A0A1Q3F0Q1 A0A182WE55 A0A182RW86 A0A0P5YUF4 A0A1A9XVX2 A0A1A9VIB6 A0A023FA40 A0A224XHZ2 A0A182H483 A0A1B6GBG3 A0A0P4VSH5 W8BRV6 A0A210Q9G8 E9HY45 A0A1W4VY12 B4MYY0 A0A0P4Y7H2 B4KJR4 B4JQX2 A0A0P5ZVT1 B3NNL6 A0A0P5U809 B4I558 B4Q7H1
PDB
1Z3H
E-value=1.07655e-42,
Score=435
Ontologies
GO
Topology
Subcellular location
Nucleus
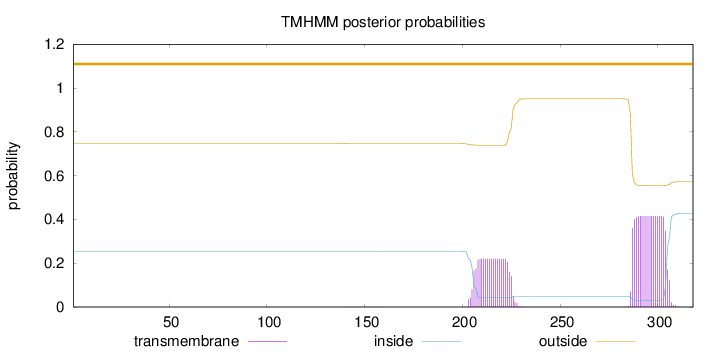
Length:
318
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
12.13617
Exp number, first 60 AAs:
0.00222
Total prob of N-in:
0.25323
outside
1 - 318
Population Genetic Test Statistics
Pi
227.654904
Theta
184.366618
Tajima's D
0.744335
CLR
0.259474
CSRT
0.585370731463427
Interpretation
Uncertain
