Gene
KWMTBOMO12123 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA004241
Annotation
PREDICTED:_exportin-2_[Papilio_xuthus]
Full name
Exportin-2
Alternative Name
Chromosome segregation 1-like protein
Importin-alpha re-exporter
Importin-alpha re-exporter
Location in the cell
PlasmaMembrane Reliability : 1.733
Sequence
CDS
ATGCTGGAGAAGTACAGCGCGTCTGGCGGCGCGGAGTGGCGCGGCAAGGACGCGGCCGTGTACCTGGTGACGTCACTGGCGTCGCGCGGCGCCACGCTGGCGGCCGGCGTGACGCGCGCCTCCCCGCTCGTGGACCTGGGACAGTTCGCCGCCAACCACGCGCTGCCCGAGCTGCAGCGGCCCAACCTGAACGAGCTGCCGGTTCTGAAGGCGGACGCCATCAAGTTCCTGATGACGTTCAGGTCGCTTCTGCCCAACGAGCTACTGCTGACCGCTTTCCCATTGCTGGTGGAGCACATCCGCGGCGCGGGCGTGGTGTGCACGTACGCGGCGTGCTGCGCGGAGAAGCTGGTGGCGGGCGGCATGGTGCGGCGCGCCGAGCTGGCCCCGCACGCCGCCGCGCTGCTCGCCGCGCTGTTCGCCGCGCTGCCCGACCACTGCACGCCCGAGCAGCACAACGAATACGTCATGAAGGCTATATTAAGGACCCTGGCGTGCCTGCAAGAGGAGGCTCTGCCGTACCTCGGCGAGGCGCTGCCGAAACTCGCCAGGATGCTGTCCGTGGTCGCCAAGAATCCTTGCAAACCTCATTTCAACCACTACCTATTCGAGACACTGTCTCTGGCTGTGTCGTTGGTGACGAAATCGAATCCAAACGCCATAGCTGCCTTCGAGGACGCCCTGTTCCCGATATTCCAAGAAATATTGCAGAACGATATACAGGAGTTCATGCCGTACGTGTTCCAAATGCTGTCGCTGCTGCTGGAGCTGCGGGGCGGGGCGGGGGGCGGCGCGGGCGAGGCGTACAGCGCGCTGCTGCCGTGCGCGGTGGCGCCCGCGCTGTGGGAGCGCGCCGCCAACGTGCGCCCGCTCGTGCGCCTGCTGTGCGCGTACGTCGCCGTCGCGCCCAACCTCGTCGTCAACTCCGGGAAACTGAACGCGATACTCGGAGTGTTTCAAAAGTTGATAGCTTCGAAAACAAACGACCATGAAGGCTTCTACTTGATACAGACTATGCTCTACAAGTTTGGACAATCTGTAATGCAGCCGTACACCAAGCAGATAATGACGCTGCTGTTCCAGCGCCTGTCCTCGTCCAAGACGACCAAGTACATCCGGGGACTGATAGCCTTCCTCGGGTTCTACGCGGTGCAGTTTGGCGCAGACCCGCTCATCGACCTCATCGACTCGGTGCAGACGAACATGTTCGCGATGTACGTGGAGCGCGTGCTGGTGTCGGAGCTGCAGCGCGTGACCGGCACGCACGAGCGCCGCGCCGCCGCCGCCGGCTGCACGCGCCTGCTCTGCACCTCCGCGCACCTGCGGGACGGCAGCCTCGCGCATCTGTGGGCGCCGCTCATGCAGGCCCTGATCCAGTTGTTCGAGCTGCCCCCGGACGAGTCCTCGCTGCCGGACGACCACTTCGTCGAGGTGGACGACGCGCCCGGCTACCAGGCGCACTACGCGCAGCTCGCCTGCGCCAAGGGACTCCTCGACGACCCGCTAGCAAAGATAGAGGATCCGAAGCGCGACCTGGTGGAGAGCTTGGAGCGCATGTCGCGCGCGCACCCCGGCGTGCTGCCCCCCCGCCTCGCCGCGCTCTCCGCCCCGCACCGCGCCGCGCTCGCCGCGTACCTCGCCGCGCACAACGTGCACCTGTCCTAA
Protein
MLEKYSASGGAEWRGKDAAVYLVTSLASRGATLAAGVTRASPLVDLGQFAANHALPELQRPNLNELPVLKADAIKFLMTFRSLLPNELLLTAFPLLVEHIRGAGVVCTYAACCAEKLVAGGMVRRAELAPHAAALLAALFAALPDHCTPEQHNEYVMKAILRTLACLQEEALPYLGEALPKLARMLSVVAKNPCKPHFNHYLFETLSLAVSLVTKSNPNAIAAFEDALFPIFQEILQNDIQEFMPYVFQMLSLLLELRGGAGGGAGEAYSALLPCAVAPALWERAANVRPLVRLLCAYVAVAPNLVVNSGKLNAILGVFQKLIASKTNDHEGFYLIQTMLYKFGQSVMQPYTKQIMTLLFQRLSSSKTTKYIRGLIAFLGFYAVQFGADPLIDLIDSVQTNMFAMYVERVLVSELQRVTGTHERRAAAAGCTRLLCTSAHLRDGSLAHLWAPLMQALIQLFELPPDESSLPDDHFVEVDDAPGYQAHYAQLACAKGLLDDPLAKIEDPKRDLVESLERMSRAHPGVLPPRLAALSAPHRAALAAYLAAHNVHLS
Summary
Description
Export receptor for importin alpha. Mediates importin-alpha re-export from the nucleus to the cytoplasm after import substrates have been released into the nucleoplasm (By similarity). Negatively regulates fluid secretion and plays a role in fluid homeostasis by downregulating cftr activity (PubMed:20933420).
Subunit
Interacts with cftr (PubMed:20933420).
Similarity
Belongs to the TRAFAC class myosin-kinesin ATPase superfamily. Myosin family.
Belongs to the XPO2/CSE1 family.
Belongs to the XPO2/CSE1 family.
Keywords
Cell membrane
Complete proteome
Cytoplasm
Membrane
Nucleus
Protein transport
Reference proteome
Transport
Feature
chain Exportin-2
Uniprot
H9J402
A0A212F558
A0A194QFC7
A0A2H1W216
A0A2D1QUE9
A0A2J7RI16
+ More
A0A1B6CNE8 A0A1B6GBG3 A0A067REG1 A0A1B6IER0 D6WTV7 W8BRV6 U4U9R7 N6U7H0 A0A2J7RI15 A0A034W6I8 A0A1Y1KAV5 A0A1S4FK97 A0A182H483 A0A0A1XDD7 A0A0K8V1J2 A0A0J7K700 Q16XY2 T1GR31 A0A195B0R1 A0A158NKK0 B0WXZ4 A0A195FBK4 A0A1Q3F0Q1 A0A151JBE9 A0A336KZ79 A0A336M2M8 F4WWC9 A0A151WRQ7 A0A0P6EQZ7 A0A084WNJ8 A0A0L0BSZ8 A0A182FVF8 W5JFU2 A0A182QT46 A0A182N2M9 A0A0P5U8H2 E2BZ56 A0A182J0K5 T1PBI4 A0A1I8NSS0 E9IFD4 K7IQA9 A0A232EYL1 A0A182WE55 A0A151IJS9 A0A0C9R0E6 A0A0P5B0W0 A0A1A9W5E5 A0A182K3X1 A0A2A3E9T1 A0A088A291 A0A0L7R791 A0A0P5ZVT1 A0A0P5U809 A0A1A9XVX2 A0A0P5PI72 A0A182PM17 A0A1J1J244 A0A0P5ZL41 A0A310SPL4 A0A1A9VIB6 A0A182U6G5 A0A182WXX2 Q7QE09 A0A182I5Z3 A0A182M1Y2 A0A182KV25 A0A182V1L9 A0A0N7ZEU1 E9HY45 A0A1B0A4R0 A0A1S3I3Z4 A0A1L8DKS7 A0A182RW86 A0A0P4Y7H2 A0A2U9BFP7 A0A182Y9N4 A0A182SBW8 A0A0K2UVJ0 Q7SZC2 A0A1B0G9Q1 B7QNR8 A0A1B0CYT2 A0A0P5ZFL7 A0A162QWR0 A0A026WED2 A0A1E1XAG0 G1KRY7 A0A023GFX0 A0A147BS71 A0A131Y3V5 A0A3N0YQG9 W5M5L4 A0A224YY17
A0A1B6CNE8 A0A1B6GBG3 A0A067REG1 A0A1B6IER0 D6WTV7 W8BRV6 U4U9R7 N6U7H0 A0A2J7RI15 A0A034W6I8 A0A1Y1KAV5 A0A1S4FK97 A0A182H483 A0A0A1XDD7 A0A0K8V1J2 A0A0J7K700 Q16XY2 T1GR31 A0A195B0R1 A0A158NKK0 B0WXZ4 A0A195FBK4 A0A1Q3F0Q1 A0A151JBE9 A0A336KZ79 A0A336M2M8 F4WWC9 A0A151WRQ7 A0A0P6EQZ7 A0A084WNJ8 A0A0L0BSZ8 A0A182FVF8 W5JFU2 A0A182QT46 A0A182N2M9 A0A0P5U8H2 E2BZ56 A0A182J0K5 T1PBI4 A0A1I8NSS0 E9IFD4 K7IQA9 A0A232EYL1 A0A182WE55 A0A151IJS9 A0A0C9R0E6 A0A0P5B0W0 A0A1A9W5E5 A0A182K3X1 A0A2A3E9T1 A0A088A291 A0A0L7R791 A0A0P5ZVT1 A0A0P5U809 A0A1A9XVX2 A0A0P5PI72 A0A182PM17 A0A1J1J244 A0A0P5ZL41 A0A310SPL4 A0A1A9VIB6 A0A182U6G5 A0A182WXX2 Q7QE09 A0A182I5Z3 A0A182M1Y2 A0A182KV25 A0A182V1L9 A0A0N7ZEU1 E9HY45 A0A1B0A4R0 A0A1S3I3Z4 A0A1L8DKS7 A0A182RW86 A0A0P4Y7H2 A0A2U9BFP7 A0A182Y9N4 A0A182SBW8 A0A0K2UVJ0 Q7SZC2 A0A1B0G9Q1 B7QNR8 A0A1B0CYT2 A0A0P5ZFL7 A0A162QWR0 A0A026WED2 A0A1E1XAG0 G1KRY7 A0A023GFX0 A0A147BS71 A0A131Y3V5 A0A3N0YQG9 W5M5L4 A0A224YY17
Pubmed
19121390
22118469
26354079
24845553
18362917
19820115
+ More
24495485 23537049 25348373 28004739 26483478 25830018 17510324 21347285 21719571 24438588 26108605 20920257 23761445 20798317 25315136 21282665 20075255 28648823 12364791 14747013 17210077 20966253 21292972 25244985 23594743 20933420 24508170 28503490 29652888 28797301
24495485 23537049 25348373 28004739 26483478 25830018 17510324 21347285 21719571 24438588 26108605 20920257 23761445 20798317 25315136 21282665 20075255 28648823 12364791 14747013 17210077 20966253 21292972 25244985 23594743 20933420 24508170 28503490 29652888 28797301
EMBL
BABH01040156
BABH01040157
BABH01040158
BABH01040159
BABH01040160
AGBW02010240
+ More
OWR48872.1 KQ459053 KPJ04187.1 ODYU01005806 SOQ47063.1 KY285054 ATP16157.1 NEVH01003506 PNF40478.1 GEDC01022433 JAS14865.1 GECZ01010007 JAS59762.1 KK852686 KDR18464.1 GECU01022326 JAS85380.1 KQ971352 EFA07339.1 GAMC01010569 JAB95986.1 KB632188 ERL89802.1 APGK01045816 KB741039 ENN74537.1 PNF40477.1 GAKP01009554 JAC49398.1 GEZM01087742 GEZM01087741 JAV58514.1 JXUM01108986 KQ565280 KXJ71133.1 GBXI01007135 GBXI01005322 JAD07157.1 JAD08970.1 GDHF01019545 JAI32769.1 LBMM01012834 KMQ85966.1 CH477530 EAT39461.1 CAQQ02190525 CAQQ02190526 KQ976692 KYM77882.1 ADTU01018869 DS232177 EDS36804.1 KQ981693 KYN37793.1 GFDL01013936 JAV21109.1 KQ979182 KYN22319.1 UFQS01001404 UFQT01001404 SSX10714.1 SSX30396.1 UFQS01000450 UFQT01000450 SSX04129.1 SSX24494.1 GL888406 EGI61471.1 KQ982805 KYQ50487.1 GDIQ01059088 JAN35649.1 ATLV01024618 KE525353 KFB51792.1 JRES01001418 KNC23108.1 ADMH02001630 ETN61689.1 AXCN02001080 GDIP01118333 JAL85381.1 GL451563 EFN79025.1 KA646122 AFP60751.1 GL762838 EFZ20732.1 NNAY01001630 OXU23377.1 KQ977294 KYN03892.1 GBYB01001470 JAG71237.1 GDIP01191317 JAJ32085.1 KZ288311 PBC28487.1 KQ414645 KOC66626.1 GDIP01038690 JAM65025.1 GDIP01118334 JAL85380.1 GDIQ01149612 JAL02114.1 CVRI01000064 CRL05526.1 GDIP01042832 JAM60883.1 KQ761976 OAD56402.1 AAAB01008848 EAA07074.1 APCN01003537 AXCM01007239 GDIP01252409 JAI70992.1 GL733109 EFX63333.1 GFDF01007018 JAV07066.1 GDIP01233710 JAI89691.1 CP026248 AWP02868.1 HACA01024355 CDW41716.1 CR559931 BC052479 BC066737 CCAG010003515 ABJB010852356 ABJB011137628 DS979863 EEC20490.1 AJVK01009334 AJVK01009335 AJVK01009336 GDIP01047185 JAM56530.1 LRGB01000248 KZS20019.1 KK107249 EZA54460.1 GFAC01002943 JAT96245.1 GBBM01003540 JAC31878.1 GEGO01001795 JAR93609.1 GEFM01002746 JAP73050.1 RJVU01031169 ROL48230.1 AHAT01025290 GFPF01007718 MAA18864.1
OWR48872.1 KQ459053 KPJ04187.1 ODYU01005806 SOQ47063.1 KY285054 ATP16157.1 NEVH01003506 PNF40478.1 GEDC01022433 JAS14865.1 GECZ01010007 JAS59762.1 KK852686 KDR18464.1 GECU01022326 JAS85380.1 KQ971352 EFA07339.1 GAMC01010569 JAB95986.1 KB632188 ERL89802.1 APGK01045816 KB741039 ENN74537.1 PNF40477.1 GAKP01009554 JAC49398.1 GEZM01087742 GEZM01087741 JAV58514.1 JXUM01108986 KQ565280 KXJ71133.1 GBXI01007135 GBXI01005322 JAD07157.1 JAD08970.1 GDHF01019545 JAI32769.1 LBMM01012834 KMQ85966.1 CH477530 EAT39461.1 CAQQ02190525 CAQQ02190526 KQ976692 KYM77882.1 ADTU01018869 DS232177 EDS36804.1 KQ981693 KYN37793.1 GFDL01013936 JAV21109.1 KQ979182 KYN22319.1 UFQS01001404 UFQT01001404 SSX10714.1 SSX30396.1 UFQS01000450 UFQT01000450 SSX04129.1 SSX24494.1 GL888406 EGI61471.1 KQ982805 KYQ50487.1 GDIQ01059088 JAN35649.1 ATLV01024618 KE525353 KFB51792.1 JRES01001418 KNC23108.1 ADMH02001630 ETN61689.1 AXCN02001080 GDIP01118333 JAL85381.1 GL451563 EFN79025.1 KA646122 AFP60751.1 GL762838 EFZ20732.1 NNAY01001630 OXU23377.1 KQ977294 KYN03892.1 GBYB01001470 JAG71237.1 GDIP01191317 JAJ32085.1 KZ288311 PBC28487.1 KQ414645 KOC66626.1 GDIP01038690 JAM65025.1 GDIP01118334 JAL85380.1 GDIQ01149612 JAL02114.1 CVRI01000064 CRL05526.1 GDIP01042832 JAM60883.1 KQ761976 OAD56402.1 AAAB01008848 EAA07074.1 APCN01003537 AXCM01007239 GDIP01252409 JAI70992.1 GL733109 EFX63333.1 GFDF01007018 JAV07066.1 GDIP01233710 JAI89691.1 CP026248 AWP02868.1 HACA01024355 CDW41716.1 CR559931 BC052479 BC066737 CCAG010003515 ABJB010852356 ABJB011137628 DS979863 EEC20490.1 AJVK01009334 AJVK01009335 AJVK01009336 GDIP01047185 JAM56530.1 LRGB01000248 KZS20019.1 KK107249 EZA54460.1 GFAC01002943 JAT96245.1 GBBM01003540 JAC31878.1 GEGO01001795 JAR93609.1 GEFM01002746 JAP73050.1 RJVU01031169 ROL48230.1 AHAT01025290 GFPF01007718 MAA18864.1
Proteomes
UP000005204
UP000007151
UP000053268
UP000235965
UP000027135
UP000007266
+ More
UP000030742 UP000019118 UP000069940 UP000249989 UP000036403 UP000008820 UP000015102 UP000078540 UP000005205 UP000002320 UP000078541 UP000078492 UP000007755 UP000075809 UP000030765 UP000037069 UP000069272 UP000000673 UP000075886 UP000075884 UP000008237 UP000075880 UP000095301 UP000095300 UP000002358 UP000215335 UP000075920 UP000078542 UP000091820 UP000075881 UP000242457 UP000005203 UP000053825 UP000092443 UP000075885 UP000183832 UP000078200 UP000075902 UP000076407 UP000007062 UP000075840 UP000075883 UP000075882 UP000075903 UP000000305 UP000092445 UP000085678 UP000075900 UP000246464 UP000076408 UP000075901 UP000000437 UP000092444 UP000001555 UP000092462 UP000076858 UP000053097 UP000001646 UP000018468
UP000030742 UP000019118 UP000069940 UP000249989 UP000036403 UP000008820 UP000015102 UP000078540 UP000005205 UP000002320 UP000078541 UP000078492 UP000007755 UP000075809 UP000030765 UP000037069 UP000069272 UP000000673 UP000075886 UP000075884 UP000008237 UP000075880 UP000095301 UP000095300 UP000002358 UP000215335 UP000075920 UP000078542 UP000091820 UP000075881 UP000242457 UP000005203 UP000053825 UP000092443 UP000075885 UP000183832 UP000078200 UP000075902 UP000076407 UP000007062 UP000075840 UP000075883 UP000075882 UP000075903 UP000000305 UP000092445 UP000085678 UP000075900 UP000246464 UP000076408 UP000075901 UP000000437 UP000092444 UP000001555 UP000092462 UP000076858 UP000053097 UP000001646 UP000018468
Pfam
Interpro
IPR011989
ARM-like
+ More
IPR013713 XPO2_central
IPR005043 XPO2_C
IPR016024 ARM-type_fold
IPR001494 Importin-beta_N
IPR029071 Ubiquitin-like_domsf
IPR027417 P-loop_NTPase
IPR000159 RA_dom
IPR002219 PE/DAG-bd
IPR001609 Myosin_head_motor_dom
IPR036961 Kinesin_motor_dom_sf
IPR008936 Rho_GTPase_activation_prot
IPR000198 RhoGAP_dom
IPR036023 MYSc_Myo9
IPR000048 IQ_motif_EF-hand-BS
IPR013713 XPO2_central
IPR005043 XPO2_C
IPR016024 ARM-type_fold
IPR001494 Importin-beta_N
IPR029071 Ubiquitin-like_domsf
IPR027417 P-loop_NTPase
IPR000159 RA_dom
IPR002219 PE/DAG-bd
IPR001609 Myosin_head_motor_dom
IPR036961 Kinesin_motor_dom_sf
IPR008936 Rho_GTPase_activation_prot
IPR000198 RhoGAP_dom
IPR036023 MYSc_Myo9
IPR000048 IQ_motif_EF-hand-BS
Gene 3D
ProteinModelPortal
H9J402
A0A212F558
A0A194QFC7
A0A2H1W216
A0A2D1QUE9
A0A2J7RI16
+ More
A0A1B6CNE8 A0A1B6GBG3 A0A067REG1 A0A1B6IER0 D6WTV7 W8BRV6 U4U9R7 N6U7H0 A0A2J7RI15 A0A034W6I8 A0A1Y1KAV5 A0A1S4FK97 A0A182H483 A0A0A1XDD7 A0A0K8V1J2 A0A0J7K700 Q16XY2 T1GR31 A0A195B0R1 A0A158NKK0 B0WXZ4 A0A195FBK4 A0A1Q3F0Q1 A0A151JBE9 A0A336KZ79 A0A336M2M8 F4WWC9 A0A151WRQ7 A0A0P6EQZ7 A0A084WNJ8 A0A0L0BSZ8 A0A182FVF8 W5JFU2 A0A182QT46 A0A182N2M9 A0A0P5U8H2 E2BZ56 A0A182J0K5 T1PBI4 A0A1I8NSS0 E9IFD4 K7IQA9 A0A232EYL1 A0A182WE55 A0A151IJS9 A0A0C9R0E6 A0A0P5B0W0 A0A1A9W5E5 A0A182K3X1 A0A2A3E9T1 A0A088A291 A0A0L7R791 A0A0P5ZVT1 A0A0P5U809 A0A1A9XVX2 A0A0P5PI72 A0A182PM17 A0A1J1J244 A0A0P5ZL41 A0A310SPL4 A0A1A9VIB6 A0A182U6G5 A0A182WXX2 Q7QE09 A0A182I5Z3 A0A182M1Y2 A0A182KV25 A0A182V1L9 A0A0N7ZEU1 E9HY45 A0A1B0A4R0 A0A1S3I3Z4 A0A1L8DKS7 A0A182RW86 A0A0P4Y7H2 A0A2U9BFP7 A0A182Y9N4 A0A182SBW8 A0A0K2UVJ0 Q7SZC2 A0A1B0G9Q1 B7QNR8 A0A1B0CYT2 A0A0P5ZFL7 A0A162QWR0 A0A026WED2 A0A1E1XAG0 G1KRY7 A0A023GFX0 A0A147BS71 A0A131Y3V5 A0A3N0YQG9 W5M5L4 A0A224YY17
A0A1B6CNE8 A0A1B6GBG3 A0A067REG1 A0A1B6IER0 D6WTV7 W8BRV6 U4U9R7 N6U7H0 A0A2J7RI15 A0A034W6I8 A0A1Y1KAV5 A0A1S4FK97 A0A182H483 A0A0A1XDD7 A0A0K8V1J2 A0A0J7K700 Q16XY2 T1GR31 A0A195B0R1 A0A158NKK0 B0WXZ4 A0A195FBK4 A0A1Q3F0Q1 A0A151JBE9 A0A336KZ79 A0A336M2M8 F4WWC9 A0A151WRQ7 A0A0P6EQZ7 A0A084WNJ8 A0A0L0BSZ8 A0A182FVF8 W5JFU2 A0A182QT46 A0A182N2M9 A0A0P5U8H2 E2BZ56 A0A182J0K5 T1PBI4 A0A1I8NSS0 E9IFD4 K7IQA9 A0A232EYL1 A0A182WE55 A0A151IJS9 A0A0C9R0E6 A0A0P5B0W0 A0A1A9W5E5 A0A182K3X1 A0A2A3E9T1 A0A088A291 A0A0L7R791 A0A0P5ZVT1 A0A0P5U809 A0A1A9XVX2 A0A0P5PI72 A0A182PM17 A0A1J1J244 A0A0P5ZL41 A0A310SPL4 A0A1A9VIB6 A0A182U6G5 A0A182WXX2 Q7QE09 A0A182I5Z3 A0A182M1Y2 A0A182KV25 A0A182V1L9 A0A0N7ZEU1 E9HY45 A0A1B0A4R0 A0A1S3I3Z4 A0A1L8DKS7 A0A182RW86 A0A0P4Y7H2 A0A2U9BFP7 A0A182Y9N4 A0A182SBW8 A0A0K2UVJ0 Q7SZC2 A0A1B0G9Q1 B7QNR8 A0A1B0CYT2 A0A0P5ZFL7 A0A162QWR0 A0A026WED2 A0A1E1XAG0 G1KRY7 A0A023GFX0 A0A147BS71 A0A131Y3V5 A0A3N0YQG9 W5M5L4 A0A224YY17
PDB
1Z3H
E-value=1.87266e-51,
Score=513
Ontologies
GO
GO:0008536
GO:0005634
GO:0006886
GO:0005623
GO:0005635
GO:0006606
GO:0006611
GO:0005829
GO:0005049
GO:0003779
GO:0046872
GO:0035556
GO:0003774
GO:0005524
GO:0016459
GO:0009925
GO:0007589
GO:0016328
GO:0016324
GO:0016021
GO:0005654
GO:0005488
GO:0005515
GO:0019001
GO:0031683
GO:0007165
GO:0005525
GO:0008033
GO:0003676
GO:0016787
Topology
Subcellular location
Nucleus
Cytoplasm Shuttles between the nucleus and the cytoplasm. With evidence from 1 publications.
Apical cell membrane Shuttles between the nucleus and the cytoplasm. With evidence from 1 publications.
Basal cell membrane Shuttles between the nucleus and the cytoplasm. With evidence from 1 publications.
Lateral cell membrane Shuttles between the nucleus and the cytoplasm. With evidence from 1 publications.
Cytoplasm Shuttles between the nucleus and the cytoplasm. With evidence from 1 publications.
Apical cell membrane Shuttles between the nucleus and the cytoplasm. With evidence from 1 publications.
Basal cell membrane Shuttles between the nucleus and the cytoplasm. With evidence from 1 publications.
Lateral cell membrane Shuttles between the nucleus and the cytoplasm. With evidence from 1 publications.
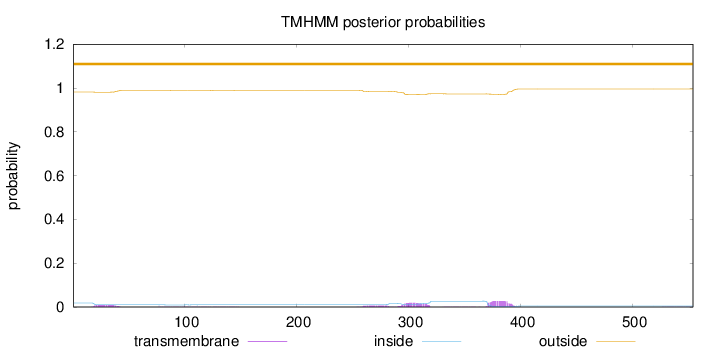
Length:
554
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.35078
Exp number, first 60 AAs:
0.18384
Total prob of N-in:
0.01846
outside
1 - 554
Population Genetic Test Statistics
Pi
181.305315
Theta
161.068498
Tajima's D
0.159213
CLR
0.221982
CSRT
0.415079246037698
Interpretation
Uncertain
