Gene
KWMTBOMO12122
Pre Gene Modal
BGIBMGA004251
Annotation
ornithine_decarboxylase_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 2.845
Sequence
CDS
ATGAAGGTTGTGGAGGAACAACGCATCAGAGTGATGGAGGGATCGTGGTCCCCGGTCAGCGTGATCCGGGAGATCGTGGAGAGCGGGGTGCAGGAGGACCCCTTCTACGTGATGGACCTCGGCGAGGTTGTCGCCCGCTACCAGCAGTGGAAGGAGCTCCTGCCCAGGGTTGAGCCGTTTTACGCTGTGAAGTGCAATGATGACAAGTTGCTGTTGACCACCCTGGCGGCTCTGGGCACTGGCTTCGATTGCGCTTCTAAAGCTGAGATCGAACTGGTCACATCTCTAGGAGTACCACCCGAGCGGATAATATTCGCGAATCCGGCTAAAATGGCGTCTCACATCCGTTACGCGTCCGCCGTCGGGGTCGACGTGATGACCTTCGACTCGGAGATTGAACTGATGAAGATCAAGCAGTACATGCCGCACGCGCAGTTGTTGATCCGCATCCGCTGCGACGCGGCCTCGGCCCAGTGCCCGCTCGGCATCAAGTTCGGTTGCGATCCCGTCACGGAAGCTCCGCGGCTGCTGAAGCTCGCCTCTGTACTCGGACTCGATGTGGCGGGCGTGTCGTTCCACGTGGGGTCGGGCGCGGCGGAGGCGGCGGTGTTCGGGCGCGGCGTGCAGCTGGCGCGCTCGCTGTTCTCGGCCGCGCGCGACCTCGGCCTGCGCCCCCGCCTGCTGGACCTGGGCGGCGGGTACCCCGGGATCGCGGCGCAGCACCACTTCATGCTAGAGATTGCGGAGGTGGTGAACGCGGCGCTGGAGAGCCATTTCCCGGAGCGCTCGGTGCGCGTGGTAGCCGAGCCCGGGCGCTACTTCGCGGCCGCCGCCTACACGCTCGCAGCCATGGTGCACTCCAAGCGGGAGATACCGGCCAAGGAGGGCGAGGACGCCGACACTCACACCATGTACTTCATCAACGACGGCGTGTACGGCTCGTTCAACTGCGTGCTGTACGACCACCAGATCGTGTCCGCTGAGCCGCTACACGATAGGGCGGATTCAGTCCTGCCTTGCTCGGTGTGGGGCCCGAGCTGTGACGCGCTGGACTGCGTCATGGAGAGCGCGCTGCTGCCGCCACTGGACGCCGGGGACTGGCTCGTGTTCCGGGACATGGGAGCGTACACCATCCCGATCGCGTCGCCCTTCAACGGGTTCCCGGTGCCGCGGGTCAGGGCCGTGGTGGACGCCGGGCTGTGGACCGGGGGCCTGCGCGAGGCCTGGCCGCCCCTGGACGAGGAGTGCCGCGCCGCGCCGGACCTCACGCCGGACCTCACGAGCGACCTCACGCCGACGCACCGCGCGGTCTTCGTCGAGTGTGCGCTCAAGTGA
Protein
MKVVEEQRIRVMEGSWSPVSVIREIVESGVQEDPFYVMDLGEVVARYQQWKELLPRVEPFYAVKCNDDKLLLTTLAALGTGFDCASKAEIELVTSLGVPPERIIFANPAKMASHIRYASAVGVDVMTFDSEIELMKIKQYMPHAQLLIRIRCDAASAQCPLGIKFGCDPVTEAPRLLKLASVLGLDVAGVSFHVGSGAAEAAVFGRGVQLARSLFSAARDLGLRPRLLDLGGGYPGIAAQHHFMLEIAEVVNAALESHFPERSVRVVAEPGRYFAAAAYTLAAMVHSKREIPAKEGEDADTHTMYFINDGVYGSFNCVLYDHQIVSAEPLHDRADSVLPCSVWGPSCDALDCVMESALLPPLDAGDWLVFRDMGAYTIPIASPFNGFPVPRVRAVVDAGLWTGGLREAWPPLDEECRAAPDLTPDLTSDLTPTHRAVFVECALK
Summary
Similarity
Belongs to the Orn/Lys/Arg decarboxylase class-II family.
Uniprot
Q1HPR3
A0A2H1W3I7
D0ENK2
A0A2A4J386
A0A194QL67
A0A3S2PLB5
+ More
A0A212F552 I4DND3 A0A2W1BUR1 A0A2P8XXQ0 A0A1B6GQR2 A0A1Y1MTA6 A0A232FHQ5 A0A154PQT1 A0A1B6CXQ9 K7IPZ1 A0A1W4X2E5 V5GTY5 A0A182FWC2 A0A2M3Z137 A0A2M3ZG90 A0A1B6GPX0 A0A1Q3EUD0 A0A1Q3EUB2 A0A1Q3FVE1 A0A1Q3EUC0 J9JQ21 A0A182H5K4 J3JZ19 A0A023ES55 U4U7P7 N6TZ37 E9HY48 A0A336K9A9 A0A2H8TR81 Q170M0 E0VIJ3 J9K5H8 A0A0B4RYF6 A0A1L8E2F1 A0A1L8E2C4 A0A161M579 A0A1B0DJ07 A0A023F2U8 A0A3R7QHL9 A0A226F1G4 A0A224XQ26 A0A0V0G351 A0A1D2MRG0 A0A1W7RA06 A0A1B6BXV7 B3S1G3 A0A3Q0JDW9 A0A1B0ACF9 A0A369S905 A7RWL7 A0A3B0JNL7 A0A1A9XUX0 D3TP90 A0A1I8P1F8 L7M043 T1IXP3 A0A1W4WAJ3 A0A1B0BUW1 A0A131YNT6 W8BLC9 A0A087TKG1 A0A3P9MD02 A0A0K8R4A6 B3MGC9 B7QC80 A0A0L0CR92 T1PM27 B5LVY4 A0A131Y2F3 A0A0A1WGE5 H2MTX0 A0A3Q2P6G7 A0A023FXE7 Q27702 A0A2B4RZV9 A0A023GK16 A0A3P9HTV6 A0A3B3DAM9 A0A034WJY6 A0A3B3BPG3 A0A315UXR3 A0A0M4E4L7 A0A1S3IDQ9 A0A0P4WJM3 A0A023FLH2 A0A3Q2ZLF8 H0VCG0 G0U8P4 A0A3Q0RVK8 T1G8U7 A0A3B3XWA6 A0A087XVR5 A0A2C9JP85
A0A212F552 I4DND3 A0A2W1BUR1 A0A2P8XXQ0 A0A1B6GQR2 A0A1Y1MTA6 A0A232FHQ5 A0A154PQT1 A0A1B6CXQ9 K7IPZ1 A0A1W4X2E5 V5GTY5 A0A182FWC2 A0A2M3Z137 A0A2M3ZG90 A0A1B6GPX0 A0A1Q3EUD0 A0A1Q3EUB2 A0A1Q3FVE1 A0A1Q3EUC0 J9JQ21 A0A182H5K4 J3JZ19 A0A023ES55 U4U7P7 N6TZ37 E9HY48 A0A336K9A9 A0A2H8TR81 Q170M0 E0VIJ3 J9K5H8 A0A0B4RYF6 A0A1L8E2F1 A0A1L8E2C4 A0A161M579 A0A1B0DJ07 A0A023F2U8 A0A3R7QHL9 A0A226F1G4 A0A224XQ26 A0A0V0G351 A0A1D2MRG0 A0A1W7RA06 A0A1B6BXV7 B3S1G3 A0A3Q0JDW9 A0A1B0ACF9 A0A369S905 A7RWL7 A0A3B0JNL7 A0A1A9XUX0 D3TP90 A0A1I8P1F8 L7M043 T1IXP3 A0A1W4WAJ3 A0A1B0BUW1 A0A131YNT6 W8BLC9 A0A087TKG1 A0A3P9MD02 A0A0K8R4A6 B3MGC9 B7QC80 A0A0L0CR92 T1PM27 B5LVY4 A0A131Y2F3 A0A0A1WGE5 H2MTX0 A0A3Q2P6G7 A0A023FXE7 Q27702 A0A2B4RZV9 A0A023GK16 A0A3P9HTV6 A0A3B3DAM9 A0A034WJY6 A0A3B3BPG3 A0A315UXR3 A0A0M4E4L7 A0A1S3IDQ9 A0A0P4WJM3 A0A023FLH2 A0A3Q2ZLF8 H0VCG0 G0U8P4 A0A3Q0RVK8 T1G8U7 A0A3B3XWA6 A0A087XVR5 A0A2C9JP85
Pubmed
19121390
26354079
22118469
22651552
28756777
29403074
+ More
28004739 28648823 20075255 26483478 22516182 24945155 23537049 21292972 17510324 20566863 25592131 25474469 27289101 18719581 30042472 17615350 20353571 25576852 26830274 24495485 17554307 17994087 26108605 18574105 25830018 25315136 29451363 25348373 29703783 21993624 22331916 23254933 15562597
28004739 28648823 20075255 26483478 22516182 24945155 23537049 21292972 17510324 20566863 25592131 25474469 27289101 18719581 30042472 17615350 20353571 25576852 26830274 24495485 17554307 17994087 26108605 18574105 25830018 25315136 29451363 25348373 29703783 21993624 22331916 23254933 15562597
EMBL
BABH01040155
DQ443339
ABF51428.1
ODYU01005806
SOQ47064.1
GQ861425
+ More
ACW82828.1 NWSH01003537 PCG66148.1 KQ459053 KPJ04186.1 RSAL01000003 RVE54717.1 AGBW02010240 OWR48871.1 AK402856 BAM19423.1 KZ149954 PZC76520.1 PYGN01001203 PSN36694.1 GECZ01005039 JAS64730.1 GEZM01023457 GEZM01023456 GEZM01023455 JAV88378.1 NNAY01000212 OXU29989.1 KQ435052 KZC14252.1 GEDC01019084 GEDC01004308 JAS18214.1 JAS32990.1 GALX01003364 JAB65102.1 GGFM01001465 MBW22216.1 GGFM01006707 MBW27458.1 GECZ01005298 JAS64471.1 GFDL01016143 JAV18902.1 GFDL01016144 JAV18901.1 GFDL01003476 JAV31569.1 GFDL01016142 JAV18903.1 ABLF02024174 JXUM01111866 KQ565544 KXJ70892.1 BT128500 AEE63457.1 JXUM01143135 GAPW01001546 KQ569540 JAC12052.1 KXJ68596.1 KB632194 ERL89919.1 APGK01055545 KB741266 ENN71527.1 GL733109 EFX63336.1 UFQS01000095 UFQT01000095 SSW99353.1 SSX19733.1 GFXV01004695 MBW16500.1 CH477472 EAT40375.1 DS235200 EEB13199.1 ABLF02037048 KJ136117 AIZ03430.1 GFDF01001193 JAV12891.1 GFDF01001194 JAV12890.1 GEMB01002453 JAS00733.1 AJVK01034626 AJVK01034627 GBBI01003353 JAC15359.1 QCYY01002764 ROT67737.1 LNIX01000001 OXA63623.1 GFTR01006183 JAW10243.1 GECL01003576 JAP02548.1 LJIJ01000677 ODM95404.1 GFAH01000407 JAV47982.1 GEDC01031414 JAS05884.1 DS985247 EDV23538.1 NOWV01000040 RDD43310.1 DS469547 EDO44188.1 OUUW01000001 SPP75169.1 EZ423242 ADD19518.1 GACK01008565 JAA56469.1 JH431658 JXJN01020939 GEDV01008801 JAP79756.1 GAMC01006958 JAB99597.1 KK115630 KFM65600.1 GADI01007818 JAA65990.1 CH902619 EDV37832.1 ABJB010042145 ABJB010120025 ABJB010141303 ABJB010236029 DS905614 EEC16452.1 JRES01000023 KNC34853.1 KA649846 AFP64475.1 EU915509 ACG76357.1 GEFM01003118 JAP72678.1 GBXI01016546 JAC97745.1 GBBL01001166 JAC26154.1 U53667 AF411043 AAA98981.1 AAL06466.1 LSMT01000251 PFX22090.1 GBBM01001141 JAC34277.1 GAKP01004869 GAKP01004867 JAC54083.1 NHOQ01002481 PWA16343.1 CP012524 ALC41028.1 GDRN01038347 JAI67258.1 GBBK01002508 JAC21974.1 AAKN02053151 HE573027 CCC53971.1 AMQM01000166 KB095811 ESO12426.1 AYCK01001308
ACW82828.1 NWSH01003537 PCG66148.1 KQ459053 KPJ04186.1 RSAL01000003 RVE54717.1 AGBW02010240 OWR48871.1 AK402856 BAM19423.1 KZ149954 PZC76520.1 PYGN01001203 PSN36694.1 GECZ01005039 JAS64730.1 GEZM01023457 GEZM01023456 GEZM01023455 JAV88378.1 NNAY01000212 OXU29989.1 KQ435052 KZC14252.1 GEDC01019084 GEDC01004308 JAS18214.1 JAS32990.1 GALX01003364 JAB65102.1 GGFM01001465 MBW22216.1 GGFM01006707 MBW27458.1 GECZ01005298 JAS64471.1 GFDL01016143 JAV18902.1 GFDL01016144 JAV18901.1 GFDL01003476 JAV31569.1 GFDL01016142 JAV18903.1 ABLF02024174 JXUM01111866 KQ565544 KXJ70892.1 BT128500 AEE63457.1 JXUM01143135 GAPW01001546 KQ569540 JAC12052.1 KXJ68596.1 KB632194 ERL89919.1 APGK01055545 KB741266 ENN71527.1 GL733109 EFX63336.1 UFQS01000095 UFQT01000095 SSW99353.1 SSX19733.1 GFXV01004695 MBW16500.1 CH477472 EAT40375.1 DS235200 EEB13199.1 ABLF02037048 KJ136117 AIZ03430.1 GFDF01001193 JAV12891.1 GFDF01001194 JAV12890.1 GEMB01002453 JAS00733.1 AJVK01034626 AJVK01034627 GBBI01003353 JAC15359.1 QCYY01002764 ROT67737.1 LNIX01000001 OXA63623.1 GFTR01006183 JAW10243.1 GECL01003576 JAP02548.1 LJIJ01000677 ODM95404.1 GFAH01000407 JAV47982.1 GEDC01031414 JAS05884.1 DS985247 EDV23538.1 NOWV01000040 RDD43310.1 DS469547 EDO44188.1 OUUW01000001 SPP75169.1 EZ423242 ADD19518.1 GACK01008565 JAA56469.1 JH431658 JXJN01020939 GEDV01008801 JAP79756.1 GAMC01006958 JAB99597.1 KK115630 KFM65600.1 GADI01007818 JAA65990.1 CH902619 EDV37832.1 ABJB010042145 ABJB010120025 ABJB010141303 ABJB010236029 DS905614 EEC16452.1 JRES01000023 KNC34853.1 KA649846 AFP64475.1 EU915509 ACG76357.1 GEFM01003118 JAP72678.1 GBXI01016546 JAC97745.1 GBBL01001166 JAC26154.1 U53667 AF411043 AAA98981.1 AAL06466.1 LSMT01000251 PFX22090.1 GBBM01001141 JAC34277.1 GAKP01004869 GAKP01004867 JAC54083.1 NHOQ01002481 PWA16343.1 CP012524 ALC41028.1 GDRN01038347 JAI67258.1 GBBK01002508 JAC21974.1 AAKN02053151 HE573027 CCC53971.1 AMQM01000166 KB095811 ESO12426.1 AYCK01001308
Proteomes
UP000005204
UP000218220
UP000053268
UP000283053
UP000007151
UP000245037
+ More
UP000215335 UP000076502 UP000002358 UP000192223 UP000069272 UP000007819 UP000069940 UP000249989 UP000030742 UP000019118 UP000000305 UP000008820 UP000009046 UP000092462 UP000283509 UP000198287 UP000094527 UP000009022 UP000079169 UP000092445 UP000253843 UP000001593 UP000268350 UP000092443 UP000095300 UP000192221 UP000092460 UP000054359 UP000265180 UP000007801 UP000001555 UP000037069 UP000001038 UP000265000 UP000095301 UP000225706 UP000265200 UP000261560 UP000092553 UP000085678 UP000264800 UP000005447 UP000261340 UP000015101 UP000261480 UP000028760 UP000076420
UP000215335 UP000076502 UP000002358 UP000192223 UP000069272 UP000007819 UP000069940 UP000249989 UP000030742 UP000019118 UP000000305 UP000008820 UP000009046 UP000092462 UP000283509 UP000198287 UP000094527 UP000009022 UP000079169 UP000092445 UP000253843 UP000001593 UP000268350 UP000092443 UP000095300 UP000192221 UP000092460 UP000054359 UP000265180 UP000007801 UP000001555 UP000037069 UP000001038 UP000265000 UP000095301 UP000225706 UP000265200 UP000261560 UP000092553 UP000085678 UP000264800 UP000005447 UP000261340 UP000015101 UP000261480 UP000028760 UP000076420
Interpro
Gene 3D
ProteinModelPortal
Q1HPR3
A0A2H1W3I7
D0ENK2
A0A2A4J386
A0A194QL67
A0A3S2PLB5
+ More
A0A212F552 I4DND3 A0A2W1BUR1 A0A2P8XXQ0 A0A1B6GQR2 A0A1Y1MTA6 A0A232FHQ5 A0A154PQT1 A0A1B6CXQ9 K7IPZ1 A0A1W4X2E5 V5GTY5 A0A182FWC2 A0A2M3Z137 A0A2M3ZG90 A0A1B6GPX0 A0A1Q3EUD0 A0A1Q3EUB2 A0A1Q3FVE1 A0A1Q3EUC0 J9JQ21 A0A182H5K4 J3JZ19 A0A023ES55 U4U7P7 N6TZ37 E9HY48 A0A336K9A9 A0A2H8TR81 Q170M0 E0VIJ3 J9K5H8 A0A0B4RYF6 A0A1L8E2F1 A0A1L8E2C4 A0A161M579 A0A1B0DJ07 A0A023F2U8 A0A3R7QHL9 A0A226F1G4 A0A224XQ26 A0A0V0G351 A0A1D2MRG0 A0A1W7RA06 A0A1B6BXV7 B3S1G3 A0A3Q0JDW9 A0A1B0ACF9 A0A369S905 A7RWL7 A0A3B0JNL7 A0A1A9XUX0 D3TP90 A0A1I8P1F8 L7M043 T1IXP3 A0A1W4WAJ3 A0A1B0BUW1 A0A131YNT6 W8BLC9 A0A087TKG1 A0A3P9MD02 A0A0K8R4A6 B3MGC9 B7QC80 A0A0L0CR92 T1PM27 B5LVY4 A0A131Y2F3 A0A0A1WGE5 H2MTX0 A0A3Q2P6G7 A0A023FXE7 Q27702 A0A2B4RZV9 A0A023GK16 A0A3P9HTV6 A0A3B3DAM9 A0A034WJY6 A0A3B3BPG3 A0A315UXR3 A0A0M4E4L7 A0A1S3IDQ9 A0A0P4WJM3 A0A023FLH2 A0A3Q2ZLF8 H0VCG0 G0U8P4 A0A3Q0RVK8 T1G8U7 A0A3B3XWA6 A0A087XVR5 A0A2C9JP85
A0A212F552 I4DND3 A0A2W1BUR1 A0A2P8XXQ0 A0A1B6GQR2 A0A1Y1MTA6 A0A232FHQ5 A0A154PQT1 A0A1B6CXQ9 K7IPZ1 A0A1W4X2E5 V5GTY5 A0A182FWC2 A0A2M3Z137 A0A2M3ZG90 A0A1B6GPX0 A0A1Q3EUD0 A0A1Q3EUB2 A0A1Q3FVE1 A0A1Q3EUC0 J9JQ21 A0A182H5K4 J3JZ19 A0A023ES55 U4U7P7 N6TZ37 E9HY48 A0A336K9A9 A0A2H8TR81 Q170M0 E0VIJ3 J9K5H8 A0A0B4RYF6 A0A1L8E2F1 A0A1L8E2C4 A0A161M579 A0A1B0DJ07 A0A023F2U8 A0A3R7QHL9 A0A226F1G4 A0A224XQ26 A0A0V0G351 A0A1D2MRG0 A0A1W7RA06 A0A1B6BXV7 B3S1G3 A0A3Q0JDW9 A0A1B0ACF9 A0A369S905 A7RWL7 A0A3B0JNL7 A0A1A9XUX0 D3TP90 A0A1I8P1F8 L7M043 T1IXP3 A0A1W4WAJ3 A0A1B0BUW1 A0A131YNT6 W8BLC9 A0A087TKG1 A0A3P9MD02 A0A0K8R4A6 B3MGC9 B7QC80 A0A0L0CR92 T1PM27 B5LVY4 A0A131Y2F3 A0A0A1WGE5 H2MTX0 A0A3Q2P6G7 A0A023FXE7 Q27702 A0A2B4RZV9 A0A023GK16 A0A3P9HTV6 A0A3B3DAM9 A0A034WJY6 A0A3B3BPG3 A0A315UXR3 A0A0M4E4L7 A0A1S3IDQ9 A0A0P4WJM3 A0A023FLH2 A0A3Q2ZLF8 H0VCG0 G0U8P4 A0A3Q0RVK8 T1G8U7 A0A3B3XWA6 A0A087XVR5 A0A2C9JP85
PDB
1NJJ
E-value=2.91194e-84,
Score=795
Ontologies
PATHWAY
GO
PANTHER
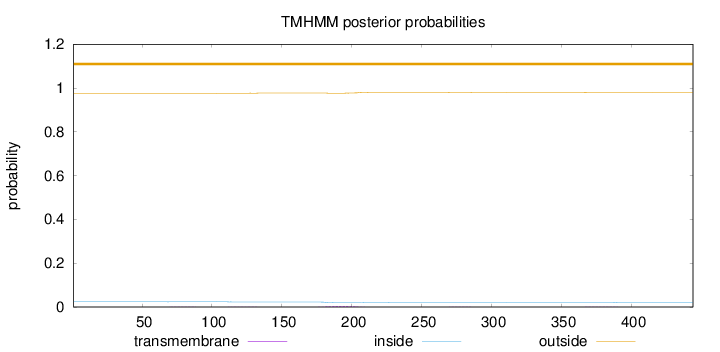
Topology
Length:
444
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.11278
Exp number, first 60 AAs:
0
Total prob of N-in:
0.02365
outside
1 - 444
Population Genetic Test Statistics
Pi
238.354392
Theta
176.626208
Tajima's D
1.12037
CLR
0
CSRT
0.693565321733913
Interpretation
Uncertain
