Gene
KWMTBOMO12119
Pre Gene Modal
BGIBMGA004243
Annotation
PREDICTED:_probable_sodium-coupled_neutral_amino_acid_transporter_6_[Amyelois_transitella]
Location in the cell
PlasmaMembrane Reliability : 4.952
Sequence
CDS
ATGAATGGAAATTGCACCCCGGAAACTCCACTCGAGGAGTCGCAGACCAAGCCCTTGGTGGTGTCTAGTCACGACCGCTACACTGTTGGCGAAACCACCGGTGGGCTCAGCGTTCTGTTTACTATCCTCTGCATAGTAGATCTCTTCGGAGTGTTTCCTGTCATAGCACTACCTAAAAGCGTGATAAGTTGCGGAATATACGGCATCCCCCTAGTCCTGTCAGTGGTTGGCCTGCAGCTATACACCGCTGCCCTGCTGGGGAGGTGCTGGCTATTGGCCAAAGAAATCACTCCTATTATCAGCGAAAAGAACAGGAGTCCATACGCAGCTGTGGCACAACTAGCGTTCGGAGATCCGGCCAGGAGGCTGGTGATCTTTCTGATCGACGCCACCGTCTTCGGCTCTGGCGTTCCAAACTTCATCTTAGCCGCCCAGAGCCTGCAGATATTCTGGTGGAAGATCTCCGGTGGTAACGTTGGCGTAACATATTGCATTTGGATGTTAGTTTTAGCCTTACTGCTCTGTCCCATAATGTGGCTCGGCTCACCCAAGGATATGAAGCCTCTCGCTCTGACGTCAGTGTTCATAGTGACCACCGTGGCGGTATCCACTTGGACGTGCATCATCCAGGACGACGTTAGCCCTGTGAGCACAGGCACCATACTGGAGTATCAGCCGCATGCTCCTGACTTCCTTATCGCTTATGGCATCTTGGCTTTTCAGTTCGATATCCATCCAATGCTGTTGACCATCCAAGTGGACATGCGCGACAGTAAGAAGGTGAACAAAGCCGTGCTCGGTGGCTTCGCCATCACCGGTTTCATGTTTACCACCACCGCTTTCCTTGTGGCCACTAGATATGGCCAAGACGTCACCAATAACATACTACAGACCATCCCGCCCTCAATACCGCTGTACCTGGTAGCGTTGTTAGTGACCCTACAGCTCTGCCTCTCCAGCGCGGTCAGCAACTCCGCACTGTTCCAGCACATCGAAGACCTGTTGCAGATCCCGAGAAATTTCTGCATACAACGCTGCTTAATCCGATCAAGCGTAGTGGCTGTCGCAGTGTTCCTAGCTGAAACAGTGCCGAGGTTCGATTTAGTGATGGGCCTCGTCGGGTCCACGCTCACGGGACCACTCATGTTCATATTCCCGCCGCTATTCTTCCTAAAACTTTGTTATTTAAAATCAAAGAAGGACATAACAGAGAGCCAAAACGCTTATTCAAATAATGCGACTAACGGTTCTTTGGACAGCCAAAAAAGCAAATTAAACAAAATCGAAACAAGCGGGTCGAGTCAACAAAATGGCGTCGTGACAGTCATCCAAAATGGCGAAAATGTCAACCCATCGTTGTACACGAAATACAAGACTTTTTCGCGCGATTACGCTGAAATGGGCAAAGAAGATGAATATACGATAAAGTGGTACGACATCGTACTGGCTCTAGTGGTGATGACCCTGGGCATCGCGGCAACAGTTATAGCAACGTATTGTAGTTGGGCACATTCGATAGAGTACGCGGAGTTCTCGCCGCCATGTCTCTTAAACGCTACCATAGCGGCTAGAAGTTTCCTGGAGAGTCAAATGGCTCTATGA
Protein
MNGNCTPETPLEESQTKPLVVSSHDRYTVGETTGGLSVLFTILCIVDLFGVFPVIALPKSVISCGIYGIPLVLSVVGLQLYTAALLGRCWLLAKEITPIISEKNRSPYAAVAQLAFGDPARRLVIFLIDATVFGSGVPNFILAAQSLQIFWWKISGGNVGVTYCIWMLVLALLLCPIMWLGSPKDMKPLALTSVFIVTTVAVSTWTCIIQDDVSPVSTGTILEYQPHAPDFLIAYGILAFQFDIHPMLLTIQVDMRDSKKVNKAVLGGFAITGFMFTTTAFLVATRYGQDVTNNILQTIPPSIPLYLVALLVTLQLCLSSAVSNSALFQHIEDLLQIPRNFCIQRCLIRSSVVAVAVFLAETVPRFDLVMGLVGSTLTGPLMFIFPPLFFLKLCYLKSKKDITESQNAYSNNATNGSLDSQKSKLNKIETSGSSQQNGVVTVIQNGENVNPSLYTKYKTFSRDYAEMGKEDEYTIKWYDIVLALVVMTLGIAATVIATYCSWAHSIEYAEFSPPCLLNATIAARSFLESQMAL
Summary
Uniprot
A0A2H1W7C6
A0A3S2LVB5
A0A2W1BXN5
A0A194QQE6
A0A194QL63
A0A212F5J9
+ More
A0A2A4JN10 A0A182Y778 A0A1B6H7I9 A0A084W1L9 A0A182GJX6 A0A1S4FNR5 Q16UF0 A0A1B0CZK0 A0A336N1P4 A0A182LFD9 B0W6F3 A0A182UFB9 A0A182UW32 Q7QBZ4 A0A336M5F5 A0A158NI94 A0A195FDN5 A0A151WR58 A0A182JHS3 A0A182Q699 A0A1J1HR61 A0A195EH59 A0A182K3C3 F4WPV8 A0A195CKR2 A0A2P8XZY1 A0A0M8ZUS8 A0A154PHQ6 A0A026WZ18 T1HBZ6 A0A0L7QQN2 A0A182W2W7 A0A310SAA7 A0A182PA47 A0A182RYG1 A0A088AA88 A0A232ET87 E2AC09 K7IYS7 W5JRB0 A0A2A3E285 A0A2P8XZR3 A0A1W4X5B8 E9IBL6 A0A2S2QHM8 A0A1I8N3G7 W8BPZ0 A0A0A1WRH3 A0A0M5J6Z8 A0A1I8P934 A0A1A9Y7D9 B3LWZ5 B4K7U6 A0A0K8VVW5 A0A023F2G5 A0A0K8UUB3 A0A1B0C0X7 A0A1B0GEY9
A0A2A4JN10 A0A182Y778 A0A1B6H7I9 A0A084W1L9 A0A182GJX6 A0A1S4FNR5 Q16UF0 A0A1B0CZK0 A0A336N1P4 A0A182LFD9 B0W6F3 A0A182UFB9 A0A182UW32 Q7QBZ4 A0A336M5F5 A0A158NI94 A0A195FDN5 A0A151WR58 A0A182JHS3 A0A182Q699 A0A1J1HR61 A0A195EH59 A0A182K3C3 F4WPV8 A0A195CKR2 A0A2P8XZY1 A0A0M8ZUS8 A0A154PHQ6 A0A026WZ18 T1HBZ6 A0A0L7QQN2 A0A182W2W7 A0A310SAA7 A0A182PA47 A0A182RYG1 A0A088AA88 A0A232ET87 E2AC09 K7IYS7 W5JRB0 A0A2A3E285 A0A2P8XZR3 A0A1W4X5B8 E9IBL6 A0A2S2QHM8 A0A1I8N3G7 W8BPZ0 A0A0A1WRH3 A0A0M5J6Z8 A0A1I8P934 A0A1A9Y7D9 B3LWZ5 B4K7U6 A0A0K8VVW5 A0A023F2G5 A0A0K8UUB3 A0A1B0C0X7 A0A1B0GEY9
Pubmed
EMBL
ODYU01006810
SOQ49005.1
RSAL01000172
RVE45114.1
KZ149954
PZC76523.1
+ More
KQ461181 KPJ07722.1 KQ459053 KPJ04181.1 AGBW02010182 OWR49008.1 NWSH01001038 PCG72974.1 GECU01037040 JAS70666.1 ATLV01019387 KE525269 KFB44113.1 JXUM01013059 JXUM01013060 KQ560367 KXJ82786.1 CH477623 EAT38147.1 AJVK01009610 UFQT01003326 SSX34873.1 DS231848 EDS36627.1 AAAB01008859 EAA07508.5 UFQT01000461 SSX24581.1 ADTU01016112 KQ981636 KYN38775.1 KQ982813 KYQ50334.1 AXCN02000218 CVRI01000015 CRK90026.1 KQ978957 KYN27199.1 GL888255 EGI63787.1 KQ977622 KYN01315.1 PYGN01001110 PSN37494.1 KQ435878 KOX70043.1 KQ434896 KZC10730.1 KK107078 QOIP01000011 EZA60389.1 RLU17194.1 ACPB03005141 KQ414786 KOC60854.1 KQ765605 OAD53868.1 NNAY01002304 OXU21580.1 GL438389 EFN69027.1 ADMH02000651 ETN65440.1 KZ288466 PBC25442.1 PSN37493.1 GL762111 EFZ22180.1 GGMS01008052 MBY77255.1 GAMC01007677 JAB98878.1 GBXI01012638 JAD01654.1 CP012526 ALC46444.1 CH902617 EDV43834.2 CH933806 EDW16467.1 GDHF01009292 JAI43022.1 GBBI01003406 JAC15306.1 GDHF01022196 JAI30118.1 JXJN01023794 CCAG010012653
KQ461181 KPJ07722.1 KQ459053 KPJ04181.1 AGBW02010182 OWR49008.1 NWSH01001038 PCG72974.1 GECU01037040 JAS70666.1 ATLV01019387 KE525269 KFB44113.1 JXUM01013059 JXUM01013060 KQ560367 KXJ82786.1 CH477623 EAT38147.1 AJVK01009610 UFQT01003326 SSX34873.1 DS231848 EDS36627.1 AAAB01008859 EAA07508.5 UFQT01000461 SSX24581.1 ADTU01016112 KQ981636 KYN38775.1 KQ982813 KYQ50334.1 AXCN02000218 CVRI01000015 CRK90026.1 KQ978957 KYN27199.1 GL888255 EGI63787.1 KQ977622 KYN01315.1 PYGN01001110 PSN37494.1 KQ435878 KOX70043.1 KQ434896 KZC10730.1 KK107078 QOIP01000011 EZA60389.1 RLU17194.1 ACPB03005141 KQ414786 KOC60854.1 KQ765605 OAD53868.1 NNAY01002304 OXU21580.1 GL438389 EFN69027.1 ADMH02000651 ETN65440.1 KZ288466 PBC25442.1 PSN37493.1 GL762111 EFZ22180.1 GGMS01008052 MBY77255.1 GAMC01007677 JAB98878.1 GBXI01012638 JAD01654.1 CP012526 ALC46444.1 CH902617 EDV43834.2 CH933806 EDW16467.1 GDHF01009292 JAI43022.1 GBBI01003406 JAC15306.1 GDHF01022196 JAI30118.1 JXJN01023794 CCAG010012653
Proteomes
UP000283053
UP000053240
UP000053268
UP000007151
UP000218220
UP000076408
+ More
UP000030765 UP000069940 UP000249989 UP000008820 UP000092462 UP000075882 UP000002320 UP000075902 UP000075903 UP000007062 UP000005205 UP000078541 UP000075809 UP000075880 UP000075886 UP000183832 UP000078492 UP000075881 UP000007755 UP000078542 UP000245037 UP000053105 UP000076502 UP000053097 UP000279307 UP000015103 UP000053825 UP000075920 UP000075885 UP000075900 UP000005203 UP000215335 UP000000311 UP000002358 UP000000673 UP000242457 UP000192223 UP000095301 UP000092553 UP000095300 UP000092443 UP000007801 UP000009192 UP000092460 UP000092444
UP000030765 UP000069940 UP000249989 UP000008820 UP000092462 UP000075882 UP000002320 UP000075902 UP000075903 UP000007062 UP000005205 UP000078541 UP000075809 UP000075880 UP000075886 UP000183832 UP000078492 UP000075881 UP000007755 UP000078542 UP000245037 UP000053105 UP000076502 UP000053097 UP000279307 UP000015103 UP000053825 UP000075920 UP000075885 UP000075900 UP000005203 UP000215335 UP000000311 UP000002358 UP000000673 UP000242457 UP000192223 UP000095301 UP000092553 UP000095300 UP000092443 UP000007801 UP000009192 UP000092460 UP000092444
SUPFAM
SSF75304
SSF75304
Gene 3D
ProteinModelPortal
A0A2H1W7C6
A0A3S2LVB5
A0A2W1BXN5
A0A194QQE6
A0A194QL63
A0A212F5J9
+ More
A0A2A4JN10 A0A182Y778 A0A1B6H7I9 A0A084W1L9 A0A182GJX6 A0A1S4FNR5 Q16UF0 A0A1B0CZK0 A0A336N1P4 A0A182LFD9 B0W6F3 A0A182UFB9 A0A182UW32 Q7QBZ4 A0A336M5F5 A0A158NI94 A0A195FDN5 A0A151WR58 A0A182JHS3 A0A182Q699 A0A1J1HR61 A0A195EH59 A0A182K3C3 F4WPV8 A0A195CKR2 A0A2P8XZY1 A0A0M8ZUS8 A0A154PHQ6 A0A026WZ18 T1HBZ6 A0A0L7QQN2 A0A182W2W7 A0A310SAA7 A0A182PA47 A0A182RYG1 A0A088AA88 A0A232ET87 E2AC09 K7IYS7 W5JRB0 A0A2A3E285 A0A2P8XZR3 A0A1W4X5B8 E9IBL6 A0A2S2QHM8 A0A1I8N3G7 W8BPZ0 A0A0A1WRH3 A0A0M5J6Z8 A0A1I8P934 A0A1A9Y7D9 B3LWZ5 B4K7U6 A0A0K8VVW5 A0A023F2G5 A0A0K8UUB3 A0A1B0C0X7 A0A1B0GEY9
A0A2A4JN10 A0A182Y778 A0A1B6H7I9 A0A084W1L9 A0A182GJX6 A0A1S4FNR5 Q16UF0 A0A1B0CZK0 A0A336N1P4 A0A182LFD9 B0W6F3 A0A182UFB9 A0A182UW32 Q7QBZ4 A0A336M5F5 A0A158NI94 A0A195FDN5 A0A151WR58 A0A182JHS3 A0A182Q699 A0A1J1HR61 A0A195EH59 A0A182K3C3 F4WPV8 A0A195CKR2 A0A2P8XZY1 A0A0M8ZUS8 A0A154PHQ6 A0A026WZ18 T1HBZ6 A0A0L7QQN2 A0A182W2W7 A0A310SAA7 A0A182PA47 A0A182RYG1 A0A088AA88 A0A232ET87 E2AC09 K7IYS7 W5JRB0 A0A2A3E285 A0A2P8XZR3 A0A1W4X5B8 E9IBL6 A0A2S2QHM8 A0A1I8N3G7 W8BPZ0 A0A0A1WRH3 A0A0M5J6Z8 A0A1I8P934 A0A1A9Y7D9 B3LWZ5 B4K7U6 A0A0K8VVW5 A0A023F2G5 A0A0K8UUB3 A0A1B0C0X7 A0A1B0GEY9
Ontologies
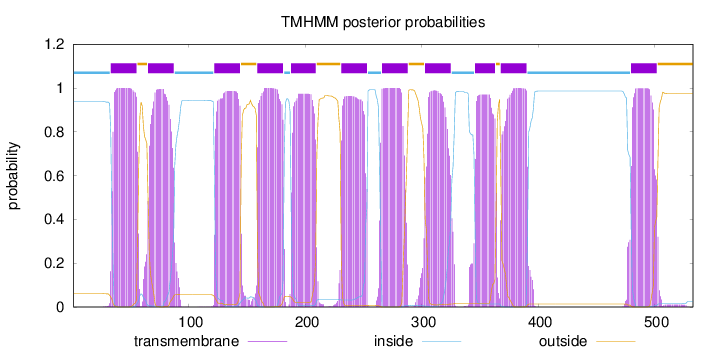
Topology
Length:
533
Number of predicted TMHs:
11
Exp number of AAs in TMHs:
239.80715
Exp number, first 60 AAs:
22.54999
Total prob of N-in:
0.93904
POSSIBLE N-term signal
sequence
inside
1 - 32
TMhelix
33 - 55
outside
56 - 64
TMhelix
65 - 87
inside
88 - 121
TMhelix
122 - 144
outside
145 - 158
TMhelix
159 - 181
inside
182 - 187
TMhelix
188 - 209
outside
210 - 230
TMhelix
231 - 253
inside
254 - 265
TMhelix
266 - 288
outside
289 - 302
TMhelix
303 - 325
inside
326 - 345
TMhelix
346 - 363
outside
364 - 367
TMhelix
368 - 390
inside
391 - 479
TMhelix
480 - 502
outside
503 - 533
Population Genetic Test Statistics
Pi
184.861847
Theta
156.589078
Tajima's D
1.187278
CLR
0.148781
CSRT
0.710664466776661
Interpretation
Uncertain
