Gene
KWMTBOMO12118 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA004249
Annotation
PREDICTED:_acyl-CoA_synthetase_family_member_3?_mitochondrial_[Bombyx_mori]
Full name
Acyl-CoA synthetase family member 3, mitochondrial
Location in the cell
Mitochondrial Reliability : 2.233
Sequence
CDS
ATGGAAATAACTTTTCGTTCGTGTCGTTTGATCAACCGGCTTTTGAGAAAACCCCTCGCTTTAACAAGTTACAGAAGTGTAGTTAATGTCGCGGCCAAAGATGAAGTGAATACCGAGCTACTGACGAGTTTCAATAAAGATATTCAAGATGGAGGTGCAGTGCCGGTGTTCCGTCGCGCTCTGCTGTTCCCGACCCGGGTCGCCTTGCAAGACGAAGTCGGGATCTACACATACGCGGGCCTCTATCAAGCCGCTGCAGCGCTGAGCACCGAAATATCTGCGCAACTTGTTGGTGAAACAGAAAGGACGATAGCTTATATGTGTGGCAACGACGCTTCGCATGTCATCGTGCAATGGGCTATATGGATGACGGGGAATATTGCCGTCCCTTTAACTCCTCTGCATCCACCGGAGATGTTAAAGTATTTCCTTACGGATAGCATGTCGACGATAGTTATATGCACCCAAGATTACGAAAAGCTTCTGCGTCCAATCGCCCTTGACCTCTCCAGGCCTTTACTGGTCACCGGGAAAGATAAAGAGATAACGGCTCAACTGTACCAGCCGAATAGCGCTTTCATGAAACCGAGGGTAGTTGATAATTTGAGCGACGTGGGTAGATCGAACTCTTGGTACGGATCGAAGGATGCGCTTATCATTTACACCAGCGGTACGACCAGTAAGCCGAAGGGCGTGGTCTGGACGCACAGCATGCTGCAGACGCAGATTGCAGCGCTGCACAGCGCCTGGCATTACAGCGGCAACGACGTCGTCTTACACACGCTGCCGTTACACCACATACACGGGCAGCTGAACTCGATGCTCGCGTCTCTGGCCGCCGGCGCCAGAATACGGATGCTGCCTTCGTTCGTATCGCACGCGGTGTGGGCCCGCCTCCTCGGCATGGGCGAGAGGGACGACTCCAAAGTGAGCGTGTTCCACGGCGTGCCGGCCATGTACATGCGGCTGGCGGCCGACCACGACAAGATGTTCGCGGACGCCCGCACCGCGGAGTACGTGCGGAACACGCTCGCCACCCGCGTGAGGCTCATGTGCGCGGGATCCGCTCCGCTACCCAGCACGCTCTTCGACAAATGGGAAAAGATATCGGGACACCGTCTCCTCGAGCGGTACGGGATGACGGAGGTGGGTATGGCGCTGAGCAACCCGTACCGTCCGCTGGAGGGTCGCACGGTGAGCTGCGTGGGGGCTCCGCTGCCGGCCGTCAGCGCCAGGATCGCGACCACCGGCACCCTCGAGCCGCTCGTCACTGTCGAGTCTCCAGCCCCCGATTTGTTGGATGGCTCCGGAGATGAATACCGGTGGGATCGGCCCAAGGTCACTGTGCACGCGCAGAGCGAGACCGAAGTCTATGAAGGGGAGCTGCTGCTGAAAGGTCCTGCAGTGTTCTCCAGATACTGGAACCGCGCGCCGAAATTAAACTCGGACGATTTCACGTCGGACGGCTGGTTCCGCACCGGCGACACCGCCTCGTACGACGGCAAGTTCAAGATCCTGGGCCGCACGTCCGTGGACATCATCAAGACGGGCGGGTACAAGGTGAGCGCCCTGCAGGTCGAGTCGGCCATCTTGGAGCACCCCGACGTCGAGGACGTGGCGGTGCTCGGCTTCGAGGATGAAAGCTTCGGGGAGATGGTCACGGCGGTGGTCGTGCTAAAAGAGAACAAAAAGCTGACCCTAAAAGACCTCAAGGACGAGGCCGGCAAGCGACTGGCGCCCTACCAGCTGCCCAAAGTCTTGCTGGTGGTGGAGCAGATGCCGAGAAACGCTATGGGCAAACTCGACAAGAAGGAGATCAGAAGACTGTACGGTGATAAACTTGGAACCAAAAAGTAG
Protein
MEITFRSCRLINRLLRKPLALTSYRSVVNVAAKDEVNTELLTSFNKDIQDGGAVPVFRRALLFPTRVALQDEVGIYTYAGLYQAAAALSTEISAQLVGETERTIAYMCGNDASHVIVQWAIWMTGNIAVPLTPLHPPEMLKYFLTDSMSTIVICTQDYEKLLRPIALDLSRPLLVTGKDKEITAQLYQPNSAFMKPRVVDNLSDVGRSNSWYGSKDALIIYTSGTTSKPKGVVWTHSMLQTQIAALHSAWHYSGNDVVLHTLPLHHIHGQLNSMLASLAAGARIRMLPSFVSHAVWARLLGMGERDDSKVSVFHGVPAMYMRLAADHDKMFADARTAEYVRNTLATRVRLMCAGSAPLPSTLFDKWEKISGHRLLERYGMTEVGMALSNPYRPLEGRTVSCVGAPLPAVSARIATTGTLEPLVTVESPAPDLLDGSGDEYRWDRPKVTVHAQSETEVYEGELLLKGPAVFSRYWNRAPKLNSDDFTSDGWFRTGDTASYDGKFKILGRTSVDIIKTGGYKVSALQVESAILEHPDVEDVAVLGFEDESFGEMVTAVVVLKENKKLTLKDLKDEAGKRLAPYQLPKVLLVVEQMPRNAMGKLDKKEIRRLYGDKLGTKK
Summary
Description
Catalyzes the initial reaction in intramitochondrial fatty acid synthesis, by activating malonate and methylmalonate, but not acetate, into their respective CoA thioester. May have some preference toward very-long-chain substrates (By similarity).
Acyl-CoA synthases catalyze the initial reaction in fatty acid metabolism, by forming a thioester with CoA. May have some preference toward very-long-chain substrates (By similarity).
Acyl-CoA synthases catalyze the initial reaction in fatty acid metabolism, by forming a thioester with CoA. May have some preference toward very-long-chain substrates (By similarity).
Similarity
Belongs to the ATP-dependent AMP-binding enzyme family.
Keywords
Alternative splicing
ATP-binding
Complete proteome
Fatty acid metabolism
Ligase
Lipid metabolism
Mitochondrion
Nucleotide-binding
Reference proteome
Transit peptide
Feature
chain Acyl-CoA synthetase family member 3, mitochondrial
splice variant In isoform 2.
splice variant In isoform 2.
Uniprot
A0A2A4JMG4
A0A2W1BSA1
A0A194QGW6
D6WR85
K7IMN4
A0A232ERZ4
+ More
A0A336MR71 A0A336LS73 J9K8Q8 A0A2J7QJN4 A0A067RBM6 A0A2S2QZR2 A0A2H8TKU3 A0A182MCY5 A0A182RSW3 N6TPN7 E0W0C8 A0A182WD24 U4UCD0 A0A182QV14 J3JTI5 A0A1S3AAI1 A0A3B4DVN8 A0A3B4DX61 W5JEL9 A0A182P1E7 A0A2M4BFL5 A0A182N3Q6 A0A2M4BFI6 A0A3B3S5L4 A0A182FTX5 A0A182YCA0 A0A2M3Z3H0 A0A093HS36 A0A0P7U821 A0A2M4BFI4 A0A182KB70 A0A2M3Z2T9 A0A1W5A4U7 A0A1U8C6D9 A0A2M3ZKQ5 A0A3Q2XT07 A0A146S230 A0A3B4AJW7 A0A3Q2PT34 A0A3Q1EB75 Q3URE1 A0A091UQW1 A0A093QZR8 U5EYT6 A0A091RYI0 A0A091PB62 A0A3B5B1G5 F1P6L8 A0A1S3KVJ8 A0A087QQ46 A0A131XXR9 V5IDV6 M4AAA2 F7DM96 I3N4B0 Q6GLK6 A0A087XLG1 G3GRS3 A0A091JYQ8 A0A3Q2ZDN8 A0A2I0LLY9 A0A3B3YF20 A0A3B5KVK5 A0A3Q7SHT5 A0A3P9MBZ9 A0A3Q1BQZ3 B2GU01 A0A1L8GEQ0 A0A3P9HY06 D3ZUX7 A0A1A8J5Z1 A0A1Q3FHS8 A0A1Q3FHS6 A0A3B3CFR2 A0A3P9QGK9 A0A091F276 A0A1A8M1P9 A0A2J8RVD0 L5LZ95 A0A1A8QF80 A0A3P8TMX2 A0A099YXD2 A0A023GE59 A0A0Q3MBU0 E9FUT1 G1CSK7 A0A3P8TWE6 A0A3B3H2K4 G3MKT8 A0A3Q0SQT1
A0A336MR71 A0A336LS73 J9K8Q8 A0A2J7QJN4 A0A067RBM6 A0A2S2QZR2 A0A2H8TKU3 A0A182MCY5 A0A182RSW3 N6TPN7 E0W0C8 A0A182WD24 U4UCD0 A0A182QV14 J3JTI5 A0A1S3AAI1 A0A3B4DVN8 A0A3B4DX61 W5JEL9 A0A182P1E7 A0A2M4BFL5 A0A182N3Q6 A0A2M4BFI6 A0A3B3S5L4 A0A182FTX5 A0A182YCA0 A0A2M3Z3H0 A0A093HS36 A0A0P7U821 A0A2M4BFI4 A0A182KB70 A0A2M3Z2T9 A0A1W5A4U7 A0A1U8C6D9 A0A2M3ZKQ5 A0A3Q2XT07 A0A146S230 A0A3B4AJW7 A0A3Q2PT34 A0A3Q1EB75 Q3URE1 A0A091UQW1 A0A093QZR8 U5EYT6 A0A091RYI0 A0A091PB62 A0A3B5B1G5 F1P6L8 A0A1S3KVJ8 A0A087QQ46 A0A131XXR9 V5IDV6 M4AAA2 F7DM96 I3N4B0 Q6GLK6 A0A087XLG1 G3GRS3 A0A091JYQ8 A0A3Q2ZDN8 A0A2I0LLY9 A0A3B3YF20 A0A3B5KVK5 A0A3Q7SHT5 A0A3P9MBZ9 A0A3Q1BQZ3 B2GU01 A0A1L8GEQ0 A0A3P9HY06 D3ZUX7 A0A1A8J5Z1 A0A1Q3FHS8 A0A1Q3FHS6 A0A3B3CFR2 A0A3P9QGK9 A0A091F276 A0A1A8M1P9 A0A2J8RVD0 L5LZ95 A0A1A8QF80 A0A3P8TMX2 A0A099YXD2 A0A023GE59 A0A0Q3MBU0 E9FUT1 G1CSK7 A0A3P8TWE6 A0A3B3H2K4 G3MKT8 A0A3Q0SQT1
EC Number
6.2.1.-
Pubmed
28756777
26354079
18362917
19820115
20075255
28648823
+ More
24845553 23537049 20566863 22516182 20920257 23761445 29240929 25244985 25463417 16141072 15489334 21183079 16341006 25765539 23542700 19892987 21804562 23371554 17554307 20431018 27762356 15057822 15632090 29451363 21292972 21841779 22216098
24845553 23537049 20566863 22516182 20920257 23761445 29240929 25244985 25463417 16141072 15489334 21183079 16341006 25765539 23542700 19892987 21804562 23371554 17554307 20431018 27762356 15057822 15632090 29451363 21292972 21841779 22216098
EMBL
NWSH01001038
PCG72976.1
KZ149954
PZC76524.1
KQ459053
KPJ04180.1
+ More
KQ971354 EFA07690.2 NNAY01002537 OXU21111.1 UFQT01001485 SSX30757.1 UFQT01000092 SSX19573.1 ABLF02025797 NEVH01013550 PNF28801.1 KK852611 KDR20268.1 GGMS01014012 MBY83215.1 GFXV01002467 MBW14272.1 AXCM01006957 APGK01059415 KB741293 ENN70246.1 DS235858 EEB19084.1 KB632046 ERL88256.1 AXCN02001102 BT126542 AEE61506.1 ADMH02001457 ETN62506.1 GGFJ01002676 MBW51817.1 GGFJ01002674 MBW51815.1 GGFM01002311 MBW23062.1 KL206429 KFV82305.1 JARO02007616 KPP63744.1 GGFJ01002675 MBW51816.1 GGFM01002086 MBW22837.1 GGFM01008358 MBW29109.1 GCES01111637 JAQ74685.1 AK141579 BC022709 KL410018 KFQ93344.1 KL224807 KFW64234.1 GANO01001820 JAB58051.1 KK936210 KFQ47249.1 KK847485 KFP88755.1 AAEX03003962 KL225802 KFM03350.1 GEFM01004781 JAP71015.1 GANP01011272 JAB73196.1 AGTP01083437 AGTP01083438 AGTP01083439 AGTP01083440 BC074473 AYCK01005499 AYCK01005500 JH000004 EGV94067.1 KK501616 KFP16651.1 AKCR02000203 PKK18431.1 AAMC01025524 AAMC01025525 AAMC01025526 AAMC01025527 AAMC01025528 AAMC01025529 BC166077 AAI66077.1 CM004473 OCT82340.1 AABR07044072 CH473972 EDL92769.1 HAED01017668 SBR04113.1 GFDL01007956 JAV27089.1 GFDL01007947 JAV27098.1 KK719441 KFO63366.1 HAEF01010480 SBR50461.1 ABGA01101904 ABGA01101905 ABGA01101906 ABGA01306119 ABGA01306120 ABGA01306121 NDHI03003646 PNJ12478.1 PNJ12479.1 PNJ12480.1 KB106264 ELK31377.1 HAEH01011542 SBR92495.1 KL886576 KGL74016.1 GBBM01003294 JAC32124.1 LMAW01002545 KQK80125.1 GL732525 EFX88868.1 JF907588 AEI69377.1 JO842489 AEO34106.1
KQ971354 EFA07690.2 NNAY01002537 OXU21111.1 UFQT01001485 SSX30757.1 UFQT01000092 SSX19573.1 ABLF02025797 NEVH01013550 PNF28801.1 KK852611 KDR20268.1 GGMS01014012 MBY83215.1 GFXV01002467 MBW14272.1 AXCM01006957 APGK01059415 KB741293 ENN70246.1 DS235858 EEB19084.1 KB632046 ERL88256.1 AXCN02001102 BT126542 AEE61506.1 ADMH02001457 ETN62506.1 GGFJ01002676 MBW51817.1 GGFJ01002674 MBW51815.1 GGFM01002311 MBW23062.1 KL206429 KFV82305.1 JARO02007616 KPP63744.1 GGFJ01002675 MBW51816.1 GGFM01002086 MBW22837.1 GGFM01008358 MBW29109.1 GCES01111637 JAQ74685.1 AK141579 BC022709 KL410018 KFQ93344.1 KL224807 KFW64234.1 GANO01001820 JAB58051.1 KK936210 KFQ47249.1 KK847485 KFP88755.1 AAEX03003962 KL225802 KFM03350.1 GEFM01004781 JAP71015.1 GANP01011272 JAB73196.1 AGTP01083437 AGTP01083438 AGTP01083439 AGTP01083440 BC074473 AYCK01005499 AYCK01005500 JH000004 EGV94067.1 KK501616 KFP16651.1 AKCR02000203 PKK18431.1 AAMC01025524 AAMC01025525 AAMC01025526 AAMC01025527 AAMC01025528 AAMC01025529 BC166077 AAI66077.1 CM004473 OCT82340.1 AABR07044072 CH473972 EDL92769.1 HAED01017668 SBR04113.1 GFDL01007956 JAV27089.1 GFDL01007947 JAV27098.1 KK719441 KFO63366.1 HAEF01010480 SBR50461.1 ABGA01101904 ABGA01101905 ABGA01101906 ABGA01306119 ABGA01306120 ABGA01306121 NDHI03003646 PNJ12478.1 PNJ12479.1 PNJ12480.1 KB106264 ELK31377.1 HAEH01011542 SBR92495.1 KL886576 KGL74016.1 GBBM01003294 JAC32124.1 LMAW01002545 KQK80125.1 GL732525 EFX88868.1 JF907588 AEI69377.1 JO842489 AEO34106.1
Proteomes
UP000218220
UP000053268
UP000007266
UP000002358
UP000215335
UP000007819
+ More
UP000235965 UP000027135 UP000075883 UP000075900 UP000019118 UP000009046 UP000075920 UP000030742 UP000075886 UP000079721 UP000261440 UP000000673 UP000075885 UP000075884 UP000261540 UP000069272 UP000076408 UP000053584 UP000034805 UP000075881 UP000192224 UP000189706 UP000264820 UP000265000 UP000261520 UP000257200 UP000000589 UP000053283 UP000054081 UP000261400 UP000002254 UP000087266 UP000053286 UP000002852 UP000002281 UP000005215 UP000028760 UP000001075 UP000053119 UP000264800 UP000053872 UP000261480 UP000261380 UP000286640 UP000265180 UP000257160 UP000008143 UP000186698 UP000265200 UP000002494 UP000261560 UP000242638 UP000052976 UP000001595 UP000265080 UP000053641 UP000051836 UP000000305 UP000001038 UP000261340
UP000235965 UP000027135 UP000075883 UP000075900 UP000019118 UP000009046 UP000075920 UP000030742 UP000075886 UP000079721 UP000261440 UP000000673 UP000075885 UP000075884 UP000261540 UP000069272 UP000076408 UP000053584 UP000034805 UP000075881 UP000192224 UP000189706 UP000264820 UP000265000 UP000261520 UP000257200 UP000000589 UP000053283 UP000054081 UP000261400 UP000002254 UP000087266 UP000053286 UP000002852 UP000002281 UP000005215 UP000028760 UP000001075 UP000053119 UP000264800 UP000053872 UP000261480 UP000261380 UP000286640 UP000265180 UP000257160 UP000008143 UP000186698 UP000265200 UP000002494 UP000261560 UP000242638 UP000052976 UP000001595 UP000265080 UP000053641 UP000051836 UP000000305 UP000001038 UP000261340
Interpro
Gene 3D
ProteinModelPortal
A0A2A4JMG4
A0A2W1BSA1
A0A194QGW6
D6WR85
K7IMN4
A0A232ERZ4
+ More
A0A336MR71 A0A336LS73 J9K8Q8 A0A2J7QJN4 A0A067RBM6 A0A2S2QZR2 A0A2H8TKU3 A0A182MCY5 A0A182RSW3 N6TPN7 E0W0C8 A0A182WD24 U4UCD0 A0A182QV14 J3JTI5 A0A1S3AAI1 A0A3B4DVN8 A0A3B4DX61 W5JEL9 A0A182P1E7 A0A2M4BFL5 A0A182N3Q6 A0A2M4BFI6 A0A3B3S5L4 A0A182FTX5 A0A182YCA0 A0A2M3Z3H0 A0A093HS36 A0A0P7U821 A0A2M4BFI4 A0A182KB70 A0A2M3Z2T9 A0A1W5A4U7 A0A1U8C6D9 A0A2M3ZKQ5 A0A3Q2XT07 A0A146S230 A0A3B4AJW7 A0A3Q2PT34 A0A3Q1EB75 Q3URE1 A0A091UQW1 A0A093QZR8 U5EYT6 A0A091RYI0 A0A091PB62 A0A3B5B1G5 F1P6L8 A0A1S3KVJ8 A0A087QQ46 A0A131XXR9 V5IDV6 M4AAA2 F7DM96 I3N4B0 Q6GLK6 A0A087XLG1 G3GRS3 A0A091JYQ8 A0A3Q2ZDN8 A0A2I0LLY9 A0A3B3YF20 A0A3B5KVK5 A0A3Q7SHT5 A0A3P9MBZ9 A0A3Q1BQZ3 B2GU01 A0A1L8GEQ0 A0A3P9HY06 D3ZUX7 A0A1A8J5Z1 A0A1Q3FHS8 A0A1Q3FHS6 A0A3B3CFR2 A0A3P9QGK9 A0A091F276 A0A1A8M1P9 A0A2J8RVD0 L5LZ95 A0A1A8QF80 A0A3P8TMX2 A0A099YXD2 A0A023GE59 A0A0Q3MBU0 E9FUT1 G1CSK7 A0A3P8TWE6 A0A3B3H2K4 G3MKT8 A0A3Q0SQT1
A0A336MR71 A0A336LS73 J9K8Q8 A0A2J7QJN4 A0A067RBM6 A0A2S2QZR2 A0A2H8TKU3 A0A182MCY5 A0A182RSW3 N6TPN7 E0W0C8 A0A182WD24 U4UCD0 A0A182QV14 J3JTI5 A0A1S3AAI1 A0A3B4DVN8 A0A3B4DX61 W5JEL9 A0A182P1E7 A0A2M4BFL5 A0A182N3Q6 A0A2M4BFI6 A0A3B3S5L4 A0A182FTX5 A0A182YCA0 A0A2M3Z3H0 A0A093HS36 A0A0P7U821 A0A2M4BFI4 A0A182KB70 A0A2M3Z2T9 A0A1W5A4U7 A0A1U8C6D9 A0A2M3ZKQ5 A0A3Q2XT07 A0A146S230 A0A3B4AJW7 A0A3Q2PT34 A0A3Q1EB75 Q3URE1 A0A091UQW1 A0A093QZR8 U5EYT6 A0A091RYI0 A0A091PB62 A0A3B5B1G5 F1P6L8 A0A1S3KVJ8 A0A087QQ46 A0A131XXR9 V5IDV6 M4AAA2 F7DM96 I3N4B0 Q6GLK6 A0A087XLG1 G3GRS3 A0A091JYQ8 A0A3Q2ZDN8 A0A2I0LLY9 A0A3B3YF20 A0A3B5KVK5 A0A3Q7SHT5 A0A3P9MBZ9 A0A3Q1BQZ3 B2GU01 A0A1L8GEQ0 A0A3P9HY06 D3ZUX7 A0A1A8J5Z1 A0A1Q3FHS8 A0A1Q3FHS6 A0A3B3CFR2 A0A3P9QGK9 A0A091F276 A0A1A8M1P9 A0A2J8RVD0 L5LZ95 A0A1A8QF80 A0A3P8TMX2 A0A099YXD2 A0A023GE59 A0A0Q3MBU0 E9FUT1 G1CSK7 A0A3P8TWE6 A0A3B3H2K4 G3MKT8 A0A3Q0SQT1
PDB
3NYR
E-value=1.78268e-52,
Score=522
Ontologies
PATHWAY
GO
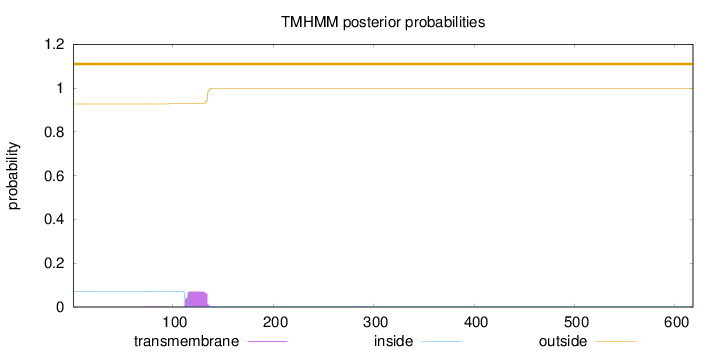
Topology
Subcellular location
Mitochondrion
Length:
618
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.56422
Exp number, first 60 AAs:
0.00021
Total prob of N-in:
0.07244
outside
1 - 618
Population Genetic Test Statistics
Pi
357.464784
Theta
169.220465
Tajima's D
0.579733
CLR
0.262483
CSRT
0.533423328833558
Interpretation
Uncertain
