Pre Gene Modal
BGIBMGA004247
Annotation
PREDICTED:_transmembrane_protein_68_isoform_X1_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.355
Sequence
CDS
ATGCGCGACATTACCCCCACTACACCAAAATCTTTGCCAACTTACGTGCCGCTTACTATATTGGAATACGTTGACACAGAATACAGCCTATGGTTGACTTGGTTCCTGACTCCTGTTATAGTGACCTTCCTCCTGCCGGCCGTCATCATAATCCTCATATACATCAGCAGTATTATATTTCACCTGTACAGATTATATAGATTAAGGGTCGTTGCCGGAGTTCAAAGCGATTGGCGTCACGCAGCGAGACTGGCTGTCTGTGCGTTGTGGGACGCTCACGGTTGGCTCTGGCACGGTTACGAGATCCGAGGCCTTGAAAATATTCCAGACGGGCCGTTCCTCGTGATCTACTATCATGGCGCTCTGCCCATAGACATGTACTACTTCATAGCCAGGATGCTGCTTTTCAAACGGAGGCACATCCACACCGTCGCCGATAGGATTCTGTTTAAAATACCGGGTTGGAAAGCGCTGCTTGAGGGCTTATGCGTTATACCTGGGACGGTCCAGACGTGCGCTTCCGTCCTGAAGGCCGGCAACCCTCTGGCCATATCGCCGGGTGGCGTTTACGAGGCCCAGTTCGGTGATCACCACTACAGGTTGAACTGGAAGTCGCGAGTCGGCTTCGCGAAAGTCGCCCTCGAAGCTAAAGTGCCGATAGTTCCGATGTTCACTCAGAACGTCCGCGAGGCGTTCCGCACGGTCGGCTGGCTCCGTAACGTCTGGTTGAGGGTGTACGCCGCCATTAGAGTGCCCCTCGCTCCAGTGTACGGCGGGTTCCCGGTCAAACTGGTCACGCACCTCGGTCCGGCCATACCTTACGACCCGGAGGCCACACCGGATCAGTTGCAGAAAAAGGTCGCCCAAGCAATAGAAGATTTAATAGAGGAACATCAAAGGATTCCAGGCAGTATATTTTTAGCGTTGGCTGAACGAATTCAAGAGCGGCCGAAACAGAAGAAATCGAGCAACACCAAGCAACTGTCAAACGGGAAATGTCAAAACGGGGACGTCAGAACGAAACAAAATGGCGTCACTGACAGCAGTCAAAGTGACAAAGCAAAGGTGTCGTAA
Protein
MRDITPTTPKSLPTYVPLTILEYVDTEYSLWLTWFLTPVIVTFLLPAVIIILIYISSIIFHLYRLYRLRVVAGVQSDWRHAARLAVCALWDAHGWLWHGYEIRGLENIPDGPFLVIYYHGALPIDMYYFIARMLLFKRRHIHTVADRILFKIPGWKALLEGLCVIPGTVQTCASVLKAGNPLAISPGGVYEAQFGDHHYRLNWKSRVGFAKVALEAKVPIVPMFTQNVREAFRTVGWLRNVWLRVYAAIRVPLAPVYGGFPVKLVTHLGPAIPYDPEATPDQLQKKVAQAIEDLIEEHQRIPGSIFLALAERIQERPKQKKSSNTKQLSNGKCQNGDVRTKQNGVTDSSQSDKAKVS
Summary
Uniprot
H9J408
A0A2H1VT13
A0A194QF57
A0A2W1BNJ9
A0A3S2TG82
A0A0L7QV44
+ More
A0A2A3ED91 A0A088A2S7 A0A154PSH2 A0A067R5L1 A0A310SKV2 A0A0N0BHP5 E2A3A6 A0A232EU01 A0A026WKH4 K7J5Z6 A0A1B6KF22 A0A0C9RVL6 A0A0C9R801 A0A1B6J306 A0A139WEC8 Q29FM9 B4HA12 B4JNW3 B4PYC8 B3NV79 A0A3B0KJ54 A0A1W4V269 B4R2S9 A0A1L8DYJ6 B4IE07 Q9VZ05 B4NEQ6 B4L3C2 B3N098 A0A0Q9WNC4 A0A1Y9H2A3 B4M1C0 A0A195BL83 A0A151IM47 A0A151WKR8 Q7PXJ9 A0A1I8JTG2 A0A182Y597 A0A182V4P9 A0A158NXL2 A0A195FH40 A0A1Y9HEK1 A0A1D2NJ54 A0A195ECI7 A0A2M4AVK2 E2B2M2 A0A2M4CPS1 A0A1B0FQ80 A0A1A9ZF46 A0A2M4CQ69 A0A2C9GSP7 A0A1A9WAT7 A0A182QJ11 A0A1A9XLX0 A0A1B0B635 A0A182S785 F4WQ17 A0A1B6GPS5 A0A1S3DIN1 A0A2S2PDY6 A0A1Y1N520 W8BJC9 A0A1I8MLT5 A0A1I8MLS6 A0A1B6CJS1 A0A2S2Q5I8 T1D4C2 J9JXS0 A0A0K8TMA8 A0A0A1X4N2 A0A0L0C2K5 A0A034VVI3 Q16M60 A0A1B0D9K2 A0A2H8TUF1 A0A1I8Q6Q6 A0A182UKE4 A0A182P2C5 W5J9M0 A0A164U3X7 A0A182F746 A0A0P5QTX1 A0A182X9Y2 U5ER09 A0A2S2QB99 A0A146M9Z9 A0A182VYU9 A0A0A9Y3N5 A0A1Q3FD57 A0A182JYD3 V5IAQ0 A0A226F2C8 E9GPS7
A0A2A3ED91 A0A088A2S7 A0A154PSH2 A0A067R5L1 A0A310SKV2 A0A0N0BHP5 E2A3A6 A0A232EU01 A0A026WKH4 K7J5Z6 A0A1B6KF22 A0A0C9RVL6 A0A0C9R801 A0A1B6J306 A0A139WEC8 Q29FM9 B4HA12 B4JNW3 B4PYC8 B3NV79 A0A3B0KJ54 A0A1W4V269 B4R2S9 A0A1L8DYJ6 B4IE07 Q9VZ05 B4NEQ6 B4L3C2 B3N098 A0A0Q9WNC4 A0A1Y9H2A3 B4M1C0 A0A195BL83 A0A151IM47 A0A151WKR8 Q7PXJ9 A0A1I8JTG2 A0A182Y597 A0A182V4P9 A0A158NXL2 A0A195FH40 A0A1Y9HEK1 A0A1D2NJ54 A0A195ECI7 A0A2M4AVK2 E2B2M2 A0A2M4CPS1 A0A1B0FQ80 A0A1A9ZF46 A0A2M4CQ69 A0A2C9GSP7 A0A1A9WAT7 A0A182QJ11 A0A1A9XLX0 A0A1B0B635 A0A182S785 F4WQ17 A0A1B6GPS5 A0A1S3DIN1 A0A2S2PDY6 A0A1Y1N520 W8BJC9 A0A1I8MLT5 A0A1I8MLS6 A0A1B6CJS1 A0A2S2Q5I8 T1D4C2 J9JXS0 A0A0K8TMA8 A0A0A1X4N2 A0A0L0C2K5 A0A034VVI3 Q16M60 A0A1B0D9K2 A0A2H8TUF1 A0A1I8Q6Q6 A0A182UKE4 A0A182P2C5 W5J9M0 A0A164U3X7 A0A182F746 A0A0P5QTX1 A0A182X9Y2 U5ER09 A0A2S2QB99 A0A146M9Z9 A0A182VYU9 A0A0A9Y3N5 A0A1Q3FD57 A0A182JYD3 V5IAQ0 A0A226F2C8 E9GPS7
Pubmed
19121390
26354079
28756777
24845553
20798317
28648823
+ More
24508170 20075255 18362917 19820115 15632085 17994087 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 18057021 12364791 14747013 17210077 25244985 21347285 27289101 21719571 28004739 24495485 25315136 24330624 26369729 25830018 26108605 25348373 17510324 20920257 23761445 26823975 25401762 21292972
24508170 20075255 18362917 19820115 15632085 17994087 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 18057021 12364791 14747013 17210077 25244985 21347285 27289101 21719571 28004739 24495485 25315136 24330624 26369729 25830018 26108605 25348373 17510324 20920257 23761445 26823975 25401762 21292972
EMBL
BABH01040142
BABH01040143
ODYU01004283
SOQ43985.1
KQ459053
KPJ04178.1
+ More
KZ149954 PZC76528.1 RSAL01000172 RVE45111.1 KQ414731 KOC62429.1 KZ288293 PBC28991.1 KQ435130 KZC14859.1 KK852686 KDR18471.1 KQ760240 OAD61311.1 KQ435750 KOX76291.1 GL436354 EFN72078.1 NNAY01002205 OXU21814.1 KK107168 EZA56126.1 GEBQ01029919 JAT10058.1 GBYB01011741 JAG81508.1 GBYB01008992 JAG78759.1 GECU01014159 JAS93547.1 KQ971354 KYB26284.1 CH379066 EAL31371.2 CH479235 EDW36680.1 CH916371 EDV92406.1 CM000162 EDX01990.1 CH954180 EDV46067.1 OUUW01000011 SPP86499.1 CM000366 EDX17628.1 GFDF01002595 JAV11489.1 CH480830 EDW45815.1 AE014298 BT016063 AAF48027.2 AAV36948.1 CH964239 EDW82225.2 CH933810 EDW07050.1 CH902640 EDV38302.1 CH940651 KRF82251.1 KRF82252.1 EDW65474.2 KRF82250.1 KQ976441 KYM86427.1 KQ977063 KYN05962.1 KQ983001 KYQ48444.1 AAAB01008987 EAA01748.5 APCN01000874 ADTU01003138 KQ981610 KYN39329.1 LJIJ01000029 ODN05145.1 KQ979074 KYN22928.1 GGFK01011480 MBW44801.1 GL445190 EFN90044.1 GGFL01003152 MBW67330.1 CCAG010006700 GGFL01003153 MBW67331.1 AXCN02000029 JXJN01008976 GL888262 EGI63715.1 GECZ01005335 JAS64434.1 GGMR01015048 MBY27667.1 GEZM01012300 JAV93001.1 GAMC01016841 JAB89714.1 GEDC01023695 JAS13603.1 GGMS01003795 MBY72998.1 GALA01001013 JAA93839.1 ABLF02040687 GDAI01002114 JAI15489.1 GBXI01008649 JAD05643.1 JRES01000984 KNC26486.1 GAKP01013409 GAKP01013408 JAC45543.1 CH477879 EAT35411.1 AJVK01028283 AJVK01028284 GFXV01006070 MBW17875.1 ADMH02001877 ETN60696.1 LRGB01001581 KZS11036.1 GDIQ01115998 JAL35728.1 GANO01003022 JAB56849.1 GGMS01005811 MBY75014.1 GDHC01021566 GDHC01002370 JAP97062.1 JAQ16259.1 GBHO01019499 GBRD01003235 JAG24105.1 JAG62586.1 GFDL01009592 JAV25453.1 GALX01000800 JAB67666.1 LNIX01000001 OXA63955.1 GL732557 EFX78435.1
KZ149954 PZC76528.1 RSAL01000172 RVE45111.1 KQ414731 KOC62429.1 KZ288293 PBC28991.1 KQ435130 KZC14859.1 KK852686 KDR18471.1 KQ760240 OAD61311.1 KQ435750 KOX76291.1 GL436354 EFN72078.1 NNAY01002205 OXU21814.1 KK107168 EZA56126.1 GEBQ01029919 JAT10058.1 GBYB01011741 JAG81508.1 GBYB01008992 JAG78759.1 GECU01014159 JAS93547.1 KQ971354 KYB26284.1 CH379066 EAL31371.2 CH479235 EDW36680.1 CH916371 EDV92406.1 CM000162 EDX01990.1 CH954180 EDV46067.1 OUUW01000011 SPP86499.1 CM000366 EDX17628.1 GFDF01002595 JAV11489.1 CH480830 EDW45815.1 AE014298 BT016063 AAF48027.2 AAV36948.1 CH964239 EDW82225.2 CH933810 EDW07050.1 CH902640 EDV38302.1 CH940651 KRF82251.1 KRF82252.1 EDW65474.2 KRF82250.1 KQ976441 KYM86427.1 KQ977063 KYN05962.1 KQ983001 KYQ48444.1 AAAB01008987 EAA01748.5 APCN01000874 ADTU01003138 KQ981610 KYN39329.1 LJIJ01000029 ODN05145.1 KQ979074 KYN22928.1 GGFK01011480 MBW44801.1 GL445190 EFN90044.1 GGFL01003152 MBW67330.1 CCAG010006700 GGFL01003153 MBW67331.1 AXCN02000029 JXJN01008976 GL888262 EGI63715.1 GECZ01005335 JAS64434.1 GGMR01015048 MBY27667.1 GEZM01012300 JAV93001.1 GAMC01016841 JAB89714.1 GEDC01023695 JAS13603.1 GGMS01003795 MBY72998.1 GALA01001013 JAA93839.1 ABLF02040687 GDAI01002114 JAI15489.1 GBXI01008649 JAD05643.1 JRES01000984 KNC26486.1 GAKP01013409 GAKP01013408 JAC45543.1 CH477879 EAT35411.1 AJVK01028283 AJVK01028284 GFXV01006070 MBW17875.1 ADMH02001877 ETN60696.1 LRGB01001581 KZS11036.1 GDIQ01115998 JAL35728.1 GANO01003022 JAB56849.1 GGMS01005811 MBY75014.1 GDHC01021566 GDHC01002370 JAP97062.1 JAQ16259.1 GBHO01019499 GBRD01003235 JAG24105.1 JAG62586.1 GFDL01009592 JAV25453.1 GALX01000800 JAB67666.1 LNIX01000001 OXA63955.1 GL732557 EFX78435.1
Proteomes
UP000005204
UP000053268
UP000283053
UP000053825
UP000242457
UP000005203
+ More
UP000076502 UP000027135 UP000053105 UP000000311 UP000215335 UP000053097 UP000002358 UP000007266 UP000001819 UP000008744 UP000001070 UP000002282 UP000008711 UP000268350 UP000192221 UP000000304 UP000001292 UP000000803 UP000007798 UP000009192 UP000007801 UP000008792 UP000075884 UP000078540 UP000078542 UP000075809 UP000007062 UP000075840 UP000076408 UP000075903 UP000005205 UP000078541 UP000075900 UP000094527 UP000078492 UP000008237 UP000092444 UP000092445 UP000075880 UP000091820 UP000075886 UP000092443 UP000092460 UP000075901 UP000007755 UP000079169 UP000095301 UP000007819 UP000037069 UP000008820 UP000092462 UP000095300 UP000075902 UP000075885 UP000000673 UP000076858 UP000069272 UP000075920 UP000075881 UP000198287 UP000000305
UP000076502 UP000027135 UP000053105 UP000000311 UP000215335 UP000053097 UP000002358 UP000007266 UP000001819 UP000008744 UP000001070 UP000002282 UP000008711 UP000268350 UP000192221 UP000000304 UP000001292 UP000000803 UP000007798 UP000009192 UP000007801 UP000008792 UP000075884 UP000078540 UP000078542 UP000075809 UP000007062 UP000075840 UP000076408 UP000075903 UP000005205 UP000078541 UP000075900 UP000094527 UP000078492 UP000008237 UP000092444 UP000092445 UP000075880 UP000091820 UP000075886 UP000092443 UP000092460 UP000075901 UP000007755 UP000079169 UP000095301 UP000007819 UP000037069 UP000008820 UP000092462 UP000095300 UP000075902 UP000075885 UP000000673 UP000076858 UP000069272 UP000075920 UP000075881 UP000198287 UP000000305
Pfam
Interpro
Gene 3D
ProteinModelPortal
H9J408
A0A2H1VT13
A0A194QF57
A0A2W1BNJ9
A0A3S2TG82
A0A0L7QV44
+ More
A0A2A3ED91 A0A088A2S7 A0A154PSH2 A0A067R5L1 A0A310SKV2 A0A0N0BHP5 E2A3A6 A0A232EU01 A0A026WKH4 K7J5Z6 A0A1B6KF22 A0A0C9RVL6 A0A0C9R801 A0A1B6J306 A0A139WEC8 Q29FM9 B4HA12 B4JNW3 B4PYC8 B3NV79 A0A3B0KJ54 A0A1W4V269 B4R2S9 A0A1L8DYJ6 B4IE07 Q9VZ05 B4NEQ6 B4L3C2 B3N098 A0A0Q9WNC4 A0A1Y9H2A3 B4M1C0 A0A195BL83 A0A151IM47 A0A151WKR8 Q7PXJ9 A0A1I8JTG2 A0A182Y597 A0A182V4P9 A0A158NXL2 A0A195FH40 A0A1Y9HEK1 A0A1D2NJ54 A0A195ECI7 A0A2M4AVK2 E2B2M2 A0A2M4CPS1 A0A1B0FQ80 A0A1A9ZF46 A0A2M4CQ69 A0A2C9GSP7 A0A1A9WAT7 A0A182QJ11 A0A1A9XLX0 A0A1B0B635 A0A182S785 F4WQ17 A0A1B6GPS5 A0A1S3DIN1 A0A2S2PDY6 A0A1Y1N520 W8BJC9 A0A1I8MLT5 A0A1I8MLS6 A0A1B6CJS1 A0A2S2Q5I8 T1D4C2 J9JXS0 A0A0K8TMA8 A0A0A1X4N2 A0A0L0C2K5 A0A034VVI3 Q16M60 A0A1B0D9K2 A0A2H8TUF1 A0A1I8Q6Q6 A0A182UKE4 A0A182P2C5 W5J9M0 A0A164U3X7 A0A182F746 A0A0P5QTX1 A0A182X9Y2 U5ER09 A0A2S2QB99 A0A146M9Z9 A0A182VYU9 A0A0A9Y3N5 A0A1Q3FD57 A0A182JYD3 V5IAQ0 A0A226F2C8 E9GPS7
A0A2A3ED91 A0A088A2S7 A0A154PSH2 A0A067R5L1 A0A310SKV2 A0A0N0BHP5 E2A3A6 A0A232EU01 A0A026WKH4 K7J5Z6 A0A1B6KF22 A0A0C9RVL6 A0A0C9R801 A0A1B6J306 A0A139WEC8 Q29FM9 B4HA12 B4JNW3 B4PYC8 B3NV79 A0A3B0KJ54 A0A1W4V269 B4R2S9 A0A1L8DYJ6 B4IE07 Q9VZ05 B4NEQ6 B4L3C2 B3N098 A0A0Q9WNC4 A0A1Y9H2A3 B4M1C0 A0A195BL83 A0A151IM47 A0A151WKR8 Q7PXJ9 A0A1I8JTG2 A0A182Y597 A0A182V4P9 A0A158NXL2 A0A195FH40 A0A1Y9HEK1 A0A1D2NJ54 A0A195ECI7 A0A2M4AVK2 E2B2M2 A0A2M4CPS1 A0A1B0FQ80 A0A1A9ZF46 A0A2M4CQ69 A0A2C9GSP7 A0A1A9WAT7 A0A182QJ11 A0A1A9XLX0 A0A1B0B635 A0A182S785 F4WQ17 A0A1B6GPS5 A0A1S3DIN1 A0A2S2PDY6 A0A1Y1N520 W8BJC9 A0A1I8MLT5 A0A1I8MLS6 A0A1B6CJS1 A0A2S2Q5I8 T1D4C2 J9JXS0 A0A0K8TMA8 A0A0A1X4N2 A0A0L0C2K5 A0A034VVI3 Q16M60 A0A1B0D9K2 A0A2H8TUF1 A0A1I8Q6Q6 A0A182UKE4 A0A182P2C5 W5J9M0 A0A164U3X7 A0A182F746 A0A0P5QTX1 A0A182X9Y2 U5ER09 A0A2S2QB99 A0A146M9Z9 A0A182VYU9 A0A0A9Y3N5 A0A1Q3FD57 A0A182JYD3 V5IAQ0 A0A226F2C8 E9GPS7
Ontologies
GO
PANTHER
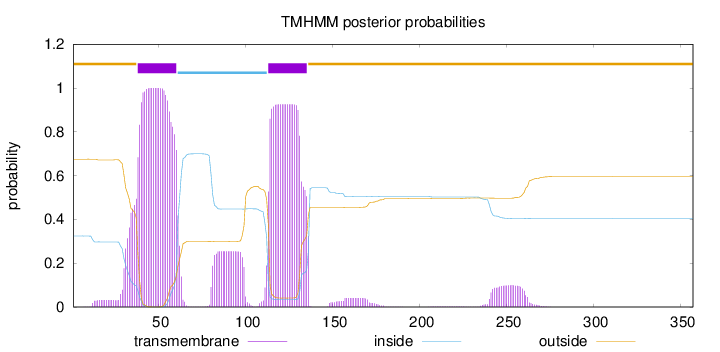
Topology
Length:
357
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
53.4732
Exp number, first 60 AAs:
24.94585
Total prob of N-in:
0.32478
POSSIBLE N-term signal
sequence
outside
1 - 37
TMhelix
38 - 60
inside
61 - 112
TMhelix
113 - 135
outside
136 - 357
Population Genetic Test Statistics
Pi
36.3218
Theta
60.149017
Tajima's D
-1.424683
CLR
0.213546
CSRT
0.0737963101844908
Interpretation
Uncertain
