Gene
KWMTBOMO12112
Annotation
reverse_transcriptase_[Bombyx_mori]
Full name
Probable RNA-directed DNA polymerase from transposon BS
Alternative Name
Reverse transcriptase
Location in the cell
Mitochondrial Reliability : 1.192 PlasmaMembrane Reliability : 1.295
Sequence
CDS
ATGGTCAGGAACGACAGGCTCTCTGCCCGTGGTGGTGGTACCGTCATTTACTATAGAAGAGCCCTGCATTGCGTCCCGCTCGATCCTCCCGCGCTCGCTAATATCGAAGCATCAGTGTGCCGAATCTCACTGACGGGACACGCGCCGATCGTTATCGCGTCCGTTTATCTTCCACCGGATAAGATCGTTCTAAGCAGTGATATCGAGGCGCTGCTCGGTATGGGGAGCTCTGTCATTCTGGCGGGCGACCTAAATTGTAAACACATCAGGTGGAACTCACACACCACAACCCCGAATGGCAGGCGGCTTGACGCGTTAGTCGATGATCTCGCCTTCGATATCGTCGCTCCGCTAACCCCGACTCACTACCCGCTAAATATCGCGCATCGCCCGGATATACTCGACATAGCGTTATTAAAAAACGTAACTCTGCGCTTACACTCGATCGAAGTAGTTTCAGAGTTAGATTCAGACCACCGTCCCGTCGTTATGAAGCTCGGTCGCGCTCCCGATTCCGTTCCCGTCACGAGGACTGTGGTGGATTGGCACACGCTGGGCATCAGCCTGGCTGAATCTGATCCACCATTGCTTCCGTTTAGTCCGGACTCTATCCCGTCTCCTCAGGATACCGCTGAAGCCATAGACATCGTAACGTCACACATCACCTCGACATTAGATAGGTCATCGAAGCAAGTTGTAGCGGAGGACTTCCTTCACCGCTTCAAATTGCCCGACGATATTAGGGAACTCCTTAGAGCTAAGAACGCCTCGATCCGTGCGTACGATAGCTCGTACTCTCAAATCGGATACGGTAGTAACCATGCCCCCCTCGAAGGCCCCTCAGGCCGACTCGCGGCGTTCGATGATGACGAAAAAGCAGAGCTGCTGGCCGATACATTGCAAACCCAGTGCACGCCCAGCACTCAATCCGTGGACCCTGTTCATGTAGAATTAGTAGACAGTGAGGTAGAACGCAGAGCCTCCTTGCCACCCTCTGATGCGTTACCACCCGTCACCCCGATGGAAGTTAAAGACTTGATCAAAGACCTACGTCCTCGCAAGGCTCCCGGTTCCGACGGTATATCCAACCGCGTTATTAAACTTCTACCCGTCCAACTCATCGTGATGTTGGCATCTATTTTCAATGCCGCTATGGCGAACTGTATCTTTCCCGCGGTGTGGAAAGAAGCGGACGTTATCGGCATACATAAACCCGGTAAACCAAAAAATCATCCGACGAGCTACCGCCCGATTAGCCTCCTCATGTCTCTAGGCAAACTGTATGAGCGTCTGCTCTACAAACGCCTCAGAGACTTCGTCTCATCCAAGGGCATTCTTATCGATGAACAATTCGGATTCCGTACAAATCACTCATGCGTTCAACAGGTGCACCGCCTCACGGAGCACATTCTTGTGGGGCTTAATCGACCAAAACCGTTATACACGGGAGCTCTCTTCTTCGACGTCGCAAAAGCGTTCGACAAAGTCTGGCACAATGGTTTGATTTTCAAACTATTCAACATGGGCGTGCCGGATAGTCTCGTGCTCATCATACGGGACTTCTTGTCGAACCGCTCTTTTCGATATCGAGTCGAGGGAACCCGCTCCTCCCCACGACCTCTCACAGCTGGAGTCCCGCAAGGCTCTGTCCTCTCACCCCTCCTATTTAGCTTATTCGTCAACGATATTCCCCGGTCGCCGCCGACCCATTTAGCTTTATTCGCCGACGACACGACTGTTTACTATTCTAGTAGAAATAAGTCCCTAATCGCGAAGAAGCTTCAGAGCGCAGCCCTAGCCCTAGGACAGTGGTTCCGAAAATGGCGCATAGACATCAACCCAGCGAAAAGTACTGCGGTGCTATTTCAGAGGGGAAGCTCCACACGGATTTCCTCCCGGATTAGGAGGAGGAATCTCACACCCCCGATTACTCTCTTTAGACAACCCATACCCTGGGCCAGGAAGGTCAAGTACCTGGGCGTTACCCTGGATGCATCGATGACATTCCGCCCGCATATAAAATCAGTCCGTGACCGTGCCGCGTTTATTCTCGGTAGACTCTACCCCATGATCTGTTAG
Protein
MVRNDRLSARGGGTVIYYRRALHCVPLDPPALANIEASVCRISLTGHAPIVIASVYLPPDKIVLSSDIEALLGMGSSVILAGDLNCKHIRWNSHTTTPNGRRLDALVDDLAFDIVAPLTPTHYPLNIAHRPDILDIALLKNVTLRLHSIEVVSELDSDHRPVVMKLGRAPDSVPVTRTVVDWHTLGISLAESDPPLLPFSPDSIPSPQDTAEAIDIVTSHITSTLDRSSKQVVAEDFLHRFKLPDDIRELLRAKNASIRAYDSSYSQIGYGSNHAPLEGPSGRLAAFDDDEKAELLADTLQTQCTPSTQSVDPVHVELVDSEVERRASLPPSDALPPVTPMEVKDLIKDLRPRKAPGSDGISNRVIKLLPVQLIVMLASIFNAAMANCIFPAVWKEADVIGIHKPGKPKNHPTSYRPISLLMSLGKLYERLLYKRLRDFVSSKGILIDEQFGFRTNHSCVQQVHRLTEHILVGLNRPKPLYTGALFFDVAKAFDKVWHNGLIFKLFNMGVPDSLVLIIRDFLSNRSFRYRVEGTRSSPRPLTAGVPQGSVLSPLLFSLFVNDIPRSPPTHLALFADDTTVYYSSRNKSLIAKKLQSAALALGQWFRKWRIDINPAKSTAVLFQRGSSTRISSRIRRRNLTPPITLFRQPIPWARKVKYLGVTLDASMTFRPHIKSVRDRAAFILGRLYPMIC
Summary
Catalytic Activity
a 2'-deoxyribonucleoside 5'-triphosphate + DNA(n) = diphosphate + DNA(n+1)
Cofactor
Mg(2+)
Mn(2+)
Mn(2+)
Keywords
Nucleotidyltransferase
RNA-directed DNA polymerase
Transferase
Transposable element
Feature
chain Probable RNA-directed DNA polymerase from transposon BS
Uniprot
Q93137
Q9BPQ0
Q9XXW0
A0A0N1PFV1
O96546
A0A1Y1NA23
+ More
V5GNJ2 D7EM17 X1WNI2 J9LC21 K7IN67 A0A023EYS8 A0A2S2QEE0 A0A2S2PPK8 X1WZH0 X1WXR6 J9JS95 J9LFC9 A0A023EZE6 J9KPF1 A0A2S2NJB5 A0A224XI99 A0A2S2N814 A0A2S2PJG2 A0A2S2PA10 A0A224XB44 X1WIX8 A0A224X7Z3 A0A2J7R7D0 A0A224X6S1 A0A2H8TF37 A0A2S2NGV8 A0A224X6T4 A0A2J7RMG0 A0A224XF24 A0A224X6U5 A0A2S2NPJ7 A0A224X6W2 A0A2S2N6Q7 A0A224X746 A0A2S2P6S3 J9KSK5 A0A2S2P698 A0A2S2Q383 A0A087UCW3 A0A0H3UA66 Q95SX7 X1WXE6 A0A0V0G5B6 A0A1Y1KFR3 A0A2S2P7B2 A0A087U3K8 J9LK81 A0A224XIB0 A0A087UGF5 A0A232ENU1 X1WTA1 A0A2S2P6C4 A0A2M4BD67 A0A2S2PBB4 A0A2H8TKI9 A0A069DX78 A0A2S2PHL7 A0A2M4BDV6 A0A2S2NKC3 A0A224X6R6 A0A2S2Q352 A0A1Q3G5J4 A0A2S2PUS3 X1XGU0
V5GNJ2 D7EM17 X1WNI2 J9LC21 K7IN67 A0A023EYS8 A0A2S2QEE0 A0A2S2PPK8 X1WZH0 X1WXR6 J9JS95 J9LFC9 A0A023EZE6 J9KPF1 A0A2S2NJB5 A0A224XI99 A0A2S2N814 A0A2S2PJG2 A0A2S2PA10 A0A224XB44 X1WIX8 A0A224X7Z3 A0A2J7R7D0 A0A224X6S1 A0A2H8TF37 A0A2S2NGV8 A0A224X6T4 A0A2J7RMG0 A0A224XF24 A0A224X6U5 A0A2S2NPJ7 A0A224X6W2 A0A2S2N6Q7 A0A224X746 A0A2S2P6S3 J9KSK5 A0A2S2P698 A0A2S2Q383 A0A087UCW3 A0A0H3UA66 Q95SX7 X1WXE6 A0A0V0G5B6 A0A1Y1KFR3 A0A2S2P7B2 A0A087U3K8 J9LK81 A0A224XIB0 A0A087UGF5 A0A232ENU1 X1WTA1 A0A2S2P6C4 A0A2M4BD67 A0A2S2PBB4 A0A2H8TKI9 A0A069DX78 A0A2S2PHL7 A0A2M4BDV6 A0A2S2NKC3 A0A224X6R6 A0A2S2Q352 A0A1Q3G5J4 A0A2S2PUS3 X1XGU0
EC Number
2.7.7.49
Pubmed
EMBL
U07847
AAA17752.1
AB055391
BAB21761.1
AB018558
BAA76304.1
+ More
KQ459265 KPJ02064.1 AF081103 AAC72921.1 GEZM01008457 JAV94772.1 GALX01005299 JAB63167.1 KQ973412 EFA12510.2 ABLF02008153 ABLF02008462 ABLF02008464 ABLF02036316 ABLF02036319 ABLF02048663 ABLF02066977 GBBI01004395 JAC14317.1 GGMS01006359 MBY75562.1 GGMR01018716 MBY31335.1 ABLF02034467 ABLF02008605 ABLF02008608 ABLF02008609 ABLF02008610 ABLF02017023 ABLF02028535 ABLF02028545 ABLF02023911 GBBI01004571 JAC14141.1 ABLF02041767 GGMR01004661 MBY17280.1 GFTR01008256 JAW08170.1 GGMR01000676 MBY13295.1 GGMR01016905 MBY29524.1 GGMR01013409 MBY26028.1 GFTR01008262 JAW08164.1 ABLF02017874 ABLF02027031 GFTR01007929 JAW08497.1 NEVH01006736 PNF36716.1 GFTR01008271 JAW08155.1 GFXV01000908 MBW12713.1 GGMR01003407 MBY16026.1 GFTR01008261 JAW08165.1 NEVH01002556 PNF42024.1 GFTR01008038 JAW08388.1 GFTR01008251 JAW08175.1 GGMR01006491 MBY19110.1 GFTR01008260 JAW08166.1 GGMR01000226 MBY12845.1 GFTR01008151 JAW08275.1 GGMR01012443 MBY25062.1 ABLF02014118 ABLF02014121 ABLF02054790 GGMR01012305 MBY24924.1 GGMS01002992 MBY72195.1 KK119261 KFM75202.1 KF280391 AIE77315.1 AY060438 AAL25477.1 ABLF02016634 GECL01003538 JAP02586.1 GEZM01084969 JAV60363.1 GGMR01012676 MBY25295.1 KK118005 KFM71947.1 ABLF02023551 ABLF02023561 GFTR01008246 JAW08180.1 KK119692 KFM76444.1 NNAY01003095 OXU19992.1 ABLF02037660 GGMR01012283 MBY24902.1 GGFJ01001858 MBW50999.1 GGMR01014053 MBY26672.1 GFXV01002840 MBW14645.1 GBGD01000389 JAC88500.1 GGMR01016296 MBY28915.1 GGFJ01001857 MBW50998.1 GGMR01005022 MBY17641.1 GFTR01008269 JAW08157.1 GGMS01002955 MBY72158.1 GFDL01000006 JAV35039.1 GGMR01020516 MBY33135.1 ABLF02055645
KQ459265 KPJ02064.1 AF081103 AAC72921.1 GEZM01008457 JAV94772.1 GALX01005299 JAB63167.1 KQ973412 EFA12510.2 ABLF02008153 ABLF02008462 ABLF02008464 ABLF02036316 ABLF02036319 ABLF02048663 ABLF02066977 GBBI01004395 JAC14317.1 GGMS01006359 MBY75562.1 GGMR01018716 MBY31335.1 ABLF02034467 ABLF02008605 ABLF02008608 ABLF02008609 ABLF02008610 ABLF02017023 ABLF02028535 ABLF02028545 ABLF02023911 GBBI01004571 JAC14141.1 ABLF02041767 GGMR01004661 MBY17280.1 GFTR01008256 JAW08170.1 GGMR01000676 MBY13295.1 GGMR01016905 MBY29524.1 GGMR01013409 MBY26028.1 GFTR01008262 JAW08164.1 ABLF02017874 ABLF02027031 GFTR01007929 JAW08497.1 NEVH01006736 PNF36716.1 GFTR01008271 JAW08155.1 GFXV01000908 MBW12713.1 GGMR01003407 MBY16026.1 GFTR01008261 JAW08165.1 NEVH01002556 PNF42024.1 GFTR01008038 JAW08388.1 GFTR01008251 JAW08175.1 GGMR01006491 MBY19110.1 GFTR01008260 JAW08166.1 GGMR01000226 MBY12845.1 GFTR01008151 JAW08275.1 GGMR01012443 MBY25062.1 ABLF02014118 ABLF02014121 ABLF02054790 GGMR01012305 MBY24924.1 GGMS01002992 MBY72195.1 KK119261 KFM75202.1 KF280391 AIE77315.1 AY060438 AAL25477.1 ABLF02016634 GECL01003538 JAP02586.1 GEZM01084969 JAV60363.1 GGMR01012676 MBY25295.1 KK118005 KFM71947.1 ABLF02023551 ABLF02023561 GFTR01008246 JAW08180.1 KK119692 KFM76444.1 NNAY01003095 OXU19992.1 ABLF02037660 GGMR01012283 MBY24902.1 GGFJ01001858 MBW50999.1 GGMR01014053 MBY26672.1 GFXV01002840 MBW14645.1 GBGD01000389 JAC88500.1 GGMR01016296 MBY28915.1 GGFJ01001857 MBW50998.1 GGMR01005022 MBY17641.1 GFTR01008269 JAW08157.1 GGMS01002955 MBY72158.1 GFDL01000006 JAV35039.1 GGMR01020516 MBY33135.1 ABLF02055645
Proteomes
Interpro
Gene 3D
ProteinModelPortal
Q93137
Q9BPQ0
Q9XXW0
A0A0N1PFV1
O96546
A0A1Y1NA23
+ More
V5GNJ2 D7EM17 X1WNI2 J9LC21 K7IN67 A0A023EYS8 A0A2S2QEE0 A0A2S2PPK8 X1WZH0 X1WXR6 J9JS95 J9LFC9 A0A023EZE6 J9KPF1 A0A2S2NJB5 A0A224XI99 A0A2S2N814 A0A2S2PJG2 A0A2S2PA10 A0A224XB44 X1WIX8 A0A224X7Z3 A0A2J7R7D0 A0A224X6S1 A0A2H8TF37 A0A2S2NGV8 A0A224X6T4 A0A2J7RMG0 A0A224XF24 A0A224X6U5 A0A2S2NPJ7 A0A224X6W2 A0A2S2N6Q7 A0A224X746 A0A2S2P6S3 J9KSK5 A0A2S2P698 A0A2S2Q383 A0A087UCW3 A0A0H3UA66 Q95SX7 X1WXE6 A0A0V0G5B6 A0A1Y1KFR3 A0A2S2P7B2 A0A087U3K8 J9LK81 A0A224XIB0 A0A087UGF5 A0A232ENU1 X1WTA1 A0A2S2P6C4 A0A2M4BD67 A0A2S2PBB4 A0A2H8TKI9 A0A069DX78 A0A2S2PHL7 A0A2M4BDV6 A0A2S2NKC3 A0A224X6R6 A0A2S2Q352 A0A1Q3G5J4 A0A2S2PUS3 X1XGU0
V5GNJ2 D7EM17 X1WNI2 J9LC21 K7IN67 A0A023EYS8 A0A2S2QEE0 A0A2S2PPK8 X1WZH0 X1WXR6 J9JS95 J9LFC9 A0A023EZE6 J9KPF1 A0A2S2NJB5 A0A224XI99 A0A2S2N814 A0A2S2PJG2 A0A2S2PA10 A0A224XB44 X1WIX8 A0A224X7Z3 A0A2J7R7D0 A0A224X6S1 A0A2H8TF37 A0A2S2NGV8 A0A224X6T4 A0A2J7RMG0 A0A224XF24 A0A224X6U5 A0A2S2NPJ7 A0A224X6W2 A0A2S2N6Q7 A0A224X746 A0A2S2P6S3 J9KSK5 A0A2S2P698 A0A2S2Q383 A0A087UCW3 A0A0H3UA66 Q95SX7 X1WXE6 A0A0V0G5B6 A0A1Y1KFR3 A0A2S2P7B2 A0A087U3K8 J9LK81 A0A224XIB0 A0A087UGF5 A0A232ENU1 X1WTA1 A0A2S2P6C4 A0A2M4BD67 A0A2S2PBB4 A0A2H8TKI9 A0A069DX78 A0A2S2PHL7 A0A2M4BDV6 A0A2S2NKC3 A0A224X6R6 A0A2S2Q352 A0A1Q3G5J4 A0A2S2PUS3 X1XGU0
Ontologies
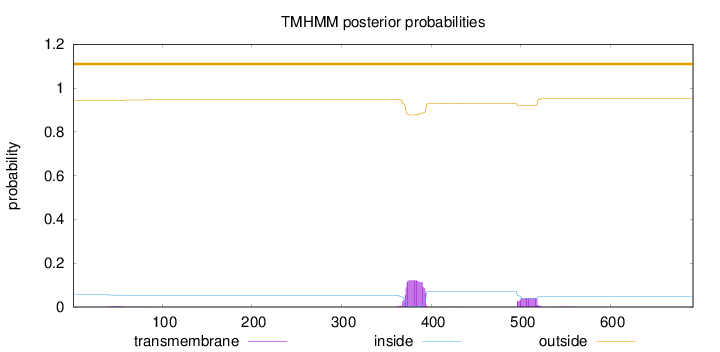
Topology
Length:
692
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
3.74234000000002
Exp number, first 60 AAs:
0.05422
Total prob of N-in:
0.05693
outside
1 - 692
Population Genetic Test Statistics
Pi
0
Theta
0
Tajima's D
0
CLR
774.224729
CSRT
0
Interpretation
Uncertain
