Gene
KWMTBOMO12111
Pre Gene Modal
BGIBMGA009686
Annotation
PREDICTED:_ceramide_glucosyltransferase_[Papilio_machaon]
Location in the cell
PlasmaMembrane Reliability : 4.043
Sequence
CDS
ATGATCCCTATAGTGTATACGGTGTATGGATTCGCGCTTTTCTTTATTGTCGCGTGGATTTGTTTGTGGCTAATACATATTATGGCGCTATCGTACTGTAAATGGAAACTTCACAGGACGGTGGACCGGTCGCCGCCCGAACAGCCTTACCCGGGCGTGTCGATCTTGAAGCCGCTCACCGGTGTAGATCCGAACCTTTTCTCGAATCTAGAGACTTTCTTTTTACTCGACTATCCGACGTACGAACTTTTGTTTTGTGTTGAGAACGAAAACGATCCGGCTATAATGTTAGTGAACAGTCTCCTACAGAAGTATCCTCAAGTGGAAGCTCGCTTGTTCGTCGGCGGACTGAATGTCGGAGTGAACCCGAAGATCAACAACATGCAACAAGGATATATCGCCGCCAAATACCCTCTTATCGTAATAAGCGACGCTGGGATCCGAATGCGAGACGATACCCTGCTGGACATGGTGCAACACTTGAAAGAGAACGTCGCGATCGTTCACCAGATGCCCTTCGCGTATGATGCCGAAGGATTTGCTGCTGTTTACGAGAAAGTATACTTCGGAACGTCACAGGCGCGAATGTACCTCGGAGCCGATTTTCTCGGCATCAATTGTCACGTCGGCATGTCGACCTTGATCCGGCGTTGCGCTATCGAGGAAGCAGGAGGACTTGCCGTGTTCGCGGACTATCTCGCTGAAGATTACTTTATGGCAAAGGAAATAGTATCGCGAGGATGGAAGATGCGGGTGGCGTCGTTGCCGGCGCTGCAGAATTCTGGGACTCGGTCTGTGGGCGCGCTGCAGGCCCGGCTCACACGGTGGGCACGGTTGCGCGTCGCCATGGTGCCGGCCACTACTCTGCTGGAGCCACTCAGCGAGTGTATGCCGTTGGGCGCAGGAGCCGCCTGGGCTGCAGGACAGCTCTTTGGAGCAGAGCCATTGGCCTTTTTCCTAGTGCATGTGTTAGTGTGGTTTCTCTCAGACTGGTTAATGTTACGGTCGGTGCAGAATGGTTCACCTCCCTTTACGAAAGTGCAATTCTTACTGGGCTGGGTTTGGAACGAGTGTTGTGCACCTTTCGTACTTGCAGTGGCGTTGCTCAGTCCTGAAATTTCATGGAGGACTCGCAGTTATCGCTTGGACTGGGGTGGCCGTGCCCGTGAACTCAGCTCCAAACTCAAATTTTAA
Protein
MIPIVYTVYGFALFFIVAWICLWLIHIMALSYCKWKLHRTVDRSPPEQPYPGVSILKPLTGVDPNLFSNLETFFLLDYPTYELLFCVENENDPAIMLVNSLLQKYPQVEARLFVGGLNVGVNPKINNMQQGYIAAKYPLIVISDAGIRMRDDTLLDMVQHLKENVAIVHQMPFAYDAEGFAAVYEKVYFGTSQARMYLGADFLGINCHVGMSTLIRRCAIEEAGGLAVFADYLAEDYFMAKEIVSRGWKMRVASLPALQNSGTRSVGALQARLTRWARLRVAMVPATTLLEPLSECMPLGAGAAWAAGQLFGAEPLAFFLVHVLVWFLSDWLMLRSVQNGSPPFTKVQFLLGWVWNECCAPFVLAVALLSPEISWRTRSYRLDWGGRARELSSKLKF
Summary
Uniprot
H9JJI5
A0A0L7KP52
A0A1E1W0G5
A0A2A4JGG7
A0A2H1WS05
A0A0N0PEF8
+ More
A0A194Q5G6 A0A212EMI0 A0A2W1BIL1 A0A2J7QMU3 A0A026WIJ9 A0A0J7NP49 E9IA29 A0A1W4X8D1 A0A0T6ATU0 A0A195FKW4 A0A154P0V2 A0A151J2Z6 A0A0L0BXI9 A0A151X0K3 B3MBH6 A0A0K8TLW6 A0A151I3S8 A0A158NVF3 A0A2A3ELG8 A0A088A139 T1HNB3 T1PI23 A0A1Y1KC89 U5ET80 A0A151I660 B4QH16 E2BSI3 A0A0C9QWI4 A0A1W4UN08 Q9W297 B4P8B1 A0A0P4VSD5 E0VZ07 B3NNC4 A0A0A9VVY6 B4LNE0 B4I7Z5 B4MP51 A0A1I8NWI7 A0A2M4AQM3 E2AW08 W5JFU9 A0A336LLW1 A0A0K8SVR4 A0A067QL28 A0A2R7WAX0 B4J8K0 A0A1L8DYS8 A0A069DSP4 A0A0M4E8T4 A0A224XE19 A0A182IVY9 A0A1Y9IWB2 A0A0V0GC91 A0A1B6D9E7 A0A182QAP4 A0A0A1XFH9 A0A034WP16 F4WR79 A0A1B6J1T7 Q17NM2 A0A3F2Z244 A0A2Y9D2X6 A0A2C9GQZ1 A0NGK2 E9HDH0 A0A0L7QU93 A0A3B0JWC1 B4GHB2 A0A182LWA6 T1KRP3 T1DI74 N6T957 A0A0P5ZIB4 Q28XN6 A0A0N8ECY0 B0XJR8 A0A0P6BUY5 B4KMV8 A0A1Q3F784 A0A1Q3F792 A0A182GNX7 A0A3S3QD11 A0A084WIW5 A0A182Y1G3 A0A182MYU6 W8C0T2 T1JHN0 A0A1J1JA77 A0A182RD16 D7EIL2 A0A1D2N9A2 A0A182P5V0 A0A0P6I429
A0A194Q5G6 A0A212EMI0 A0A2W1BIL1 A0A2J7QMU3 A0A026WIJ9 A0A0J7NP49 E9IA29 A0A1W4X8D1 A0A0T6ATU0 A0A195FKW4 A0A154P0V2 A0A151J2Z6 A0A0L0BXI9 A0A151X0K3 B3MBH6 A0A0K8TLW6 A0A151I3S8 A0A158NVF3 A0A2A3ELG8 A0A088A139 T1HNB3 T1PI23 A0A1Y1KC89 U5ET80 A0A151I660 B4QH16 E2BSI3 A0A0C9QWI4 A0A1W4UN08 Q9W297 B4P8B1 A0A0P4VSD5 E0VZ07 B3NNC4 A0A0A9VVY6 B4LNE0 B4I7Z5 B4MP51 A0A1I8NWI7 A0A2M4AQM3 E2AW08 W5JFU9 A0A336LLW1 A0A0K8SVR4 A0A067QL28 A0A2R7WAX0 B4J8K0 A0A1L8DYS8 A0A069DSP4 A0A0M4E8T4 A0A224XE19 A0A182IVY9 A0A1Y9IWB2 A0A0V0GC91 A0A1B6D9E7 A0A182QAP4 A0A0A1XFH9 A0A034WP16 F4WR79 A0A1B6J1T7 Q17NM2 A0A3F2Z244 A0A2Y9D2X6 A0A2C9GQZ1 A0NGK2 E9HDH0 A0A0L7QU93 A0A3B0JWC1 B4GHB2 A0A182LWA6 T1KRP3 T1DI74 N6T957 A0A0P5ZIB4 Q28XN6 A0A0N8ECY0 B0XJR8 A0A0P6BUY5 B4KMV8 A0A1Q3F784 A0A1Q3F792 A0A182GNX7 A0A3S3QD11 A0A084WIW5 A0A182Y1G3 A0A182MYU6 W8C0T2 T1JHN0 A0A1J1JA77 A0A182RD16 D7EIL2 A0A1D2N9A2 A0A182P5V0 A0A0P6I429
Pubmed
19121390
26227816
26354079
22118469
28756777
24508170
+ More
30249741 21282665 26108605 17994087 26369729 21347285 25315136 28004739 22936249 20798317 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 27129103 20566863 25401762 26823975 20920257 23761445 24845553 26334808 25830018 25348373 21719571 17510324 12364791 14747013 17210077 21292972 24330624 23537049 15632085 26483478 24438588 25244985 24495485 18362917 19820115 27289101
30249741 21282665 26108605 17994087 26369729 21347285 25315136 28004739 22936249 20798317 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 27129103 20566863 25401762 26823975 20920257 23761445 24845553 26334808 25830018 25348373 21719571 17510324 12364791 14747013 17210077 21292972 24330624 23537049 15632085 26483478 24438588 25244985 24495485 18362917 19820115 27289101
EMBL
BABH01017505
JTDY01007617
KOB65072.1
GDQN01010635
JAT80419.1
NWSH01001664
+ More
PCG70482.1 ODYU01010584 SOQ55797.1 KQ459838 KPJ19728.1 KQ459460 KPJ00792.1 AGBW02013827 OWR42693.1 KZ150305 PZC71513.1 NEVH01013202 PNF29910.1 KK107234 QOIP01000013 EZA54929.1 RLU15258.1 LBMM01002867 KMQ94270.1 GL761952 EFZ22574.1 LJIG01022815 KRT78575.1 KQ981512 KYN40902.1 KQ434792 KZC05452.1 KQ980313 KYN16699.1 JRES01001179 KNC24752.1 KQ982617 KYQ53769.1 CH902619 EDV37107.1 GDAI01002236 JAI15367.1 KQ976478 KYM83801.1 ADTU01027126 KZ288215 PBC32653.1 ACPB03020356 KA648334 AFP62963.1 GEZM01088569 JAV58098.1 GANO01002046 JAB57825.1 KQ978503 KYM93408.1 CM000362 CM002911 EDX08162.1 KMY95701.1 GL450237 EFN81347.1 GBYB01008104 JAG77871.1 AE013599 AY118335 AAF46795.1 AAM48364.1 CM000158 EDW91151.1 GDKW01001753 JAI54842.1 DS235848 EEB18613.1 CH954179 EDV55548.1 GBHO01044243 GBHO01044241 GDHC01000152 JAF99360.1 JAF99362.1 JAQ18477.1 CH940648 EDW61092.1 CH480824 EDW56720.1 CH963848 EDW73890.1 GGFK01009774 MBW43095.1 GL443213 EFN62430.1 ADMH02001278 ETN63252.1 UFQT01000048 SSX18966.1 GBRD01008578 GBRD01008575 JAG57246.1 KK853224 KDR09706.1 KK854557 PTY16832.1 CH916367 EDW01267.1 GFDF01002659 JAV11425.1 GBGD01001771 JAC87118.1 CP012524 ALC41196.1 GFTR01005729 JAW10697.1 GECL01000526 JAP05598.1 GEDC01014991 GEDC01008797 JAS22307.1 JAS28501.1 AXCN02001027 GBXI01004133 JAD10159.1 GAKP01001663 GAKP01001662 GAKP01001661 JAC57291.1 GL888284 EGI63245.1 GECU01014568 JAS93138.1 CH477198 EAT48292.1 APCN01003310 AAAB01008986 EAU75755.1 GL732624 EFX70139.1 KQ414735 KOC62213.1 OUUW01000001 SPP75378.1 CH479183 EDW35882.1 AXCM01001414 CAEY01000417 GALA01001101 JAA93751.1 APGK01047232 KB741077 KB631792 ENN74253.1 ERL86120.1 GDIP01056704 JAM47011.1 CM000071 EAL26280.1 GDIQ01039028 LRGB01000141 JAN55709.1 KZS20635.1 DS233582 EDS30631.1 GDIP01009432 JAM94283.1 CH933808 EDW09880.1 GFDL01011621 JAV23424.1 GFDL01011646 JAV23399.1 JXUM01077332 KQ563002 KXJ74655.1 NCKU01003556 NCKU01002258 RWS07297.1 RWS09995.1 ATLV01023950 KE525347 KFB50159.1 GAMC01003847 GAMC01003846 GAMC01003842 JAC02709.1 JH431789 CVRI01000075 CRL08446.1 KQ971307 EFA12048.1 LJIJ01000144 ODN01675.1 GDIQ01009638 JAN85099.1
PCG70482.1 ODYU01010584 SOQ55797.1 KQ459838 KPJ19728.1 KQ459460 KPJ00792.1 AGBW02013827 OWR42693.1 KZ150305 PZC71513.1 NEVH01013202 PNF29910.1 KK107234 QOIP01000013 EZA54929.1 RLU15258.1 LBMM01002867 KMQ94270.1 GL761952 EFZ22574.1 LJIG01022815 KRT78575.1 KQ981512 KYN40902.1 KQ434792 KZC05452.1 KQ980313 KYN16699.1 JRES01001179 KNC24752.1 KQ982617 KYQ53769.1 CH902619 EDV37107.1 GDAI01002236 JAI15367.1 KQ976478 KYM83801.1 ADTU01027126 KZ288215 PBC32653.1 ACPB03020356 KA648334 AFP62963.1 GEZM01088569 JAV58098.1 GANO01002046 JAB57825.1 KQ978503 KYM93408.1 CM000362 CM002911 EDX08162.1 KMY95701.1 GL450237 EFN81347.1 GBYB01008104 JAG77871.1 AE013599 AY118335 AAF46795.1 AAM48364.1 CM000158 EDW91151.1 GDKW01001753 JAI54842.1 DS235848 EEB18613.1 CH954179 EDV55548.1 GBHO01044243 GBHO01044241 GDHC01000152 JAF99360.1 JAF99362.1 JAQ18477.1 CH940648 EDW61092.1 CH480824 EDW56720.1 CH963848 EDW73890.1 GGFK01009774 MBW43095.1 GL443213 EFN62430.1 ADMH02001278 ETN63252.1 UFQT01000048 SSX18966.1 GBRD01008578 GBRD01008575 JAG57246.1 KK853224 KDR09706.1 KK854557 PTY16832.1 CH916367 EDW01267.1 GFDF01002659 JAV11425.1 GBGD01001771 JAC87118.1 CP012524 ALC41196.1 GFTR01005729 JAW10697.1 GECL01000526 JAP05598.1 GEDC01014991 GEDC01008797 JAS22307.1 JAS28501.1 AXCN02001027 GBXI01004133 JAD10159.1 GAKP01001663 GAKP01001662 GAKP01001661 JAC57291.1 GL888284 EGI63245.1 GECU01014568 JAS93138.1 CH477198 EAT48292.1 APCN01003310 AAAB01008986 EAU75755.1 GL732624 EFX70139.1 KQ414735 KOC62213.1 OUUW01000001 SPP75378.1 CH479183 EDW35882.1 AXCM01001414 CAEY01000417 GALA01001101 JAA93751.1 APGK01047232 KB741077 KB631792 ENN74253.1 ERL86120.1 GDIP01056704 JAM47011.1 CM000071 EAL26280.1 GDIQ01039028 LRGB01000141 JAN55709.1 KZS20635.1 DS233582 EDS30631.1 GDIP01009432 JAM94283.1 CH933808 EDW09880.1 GFDL01011621 JAV23424.1 GFDL01011646 JAV23399.1 JXUM01077332 KQ563002 KXJ74655.1 NCKU01003556 NCKU01002258 RWS07297.1 RWS09995.1 ATLV01023950 KE525347 KFB50159.1 GAMC01003847 GAMC01003846 GAMC01003842 JAC02709.1 JH431789 CVRI01000075 CRL08446.1 KQ971307 EFA12048.1 LJIJ01000144 ODN01675.1 GDIQ01009638 JAN85099.1
Proteomes
UP000005204
UP000037510
UP000218220
UP000053240
UP000053268
UP000007151
+ More
UP000235965 UP000053097 UP000279307 UP000036403 UP000192223 UP000078541 UP000076502 UP000078492 UP000037069 UP000075809 UP000007801 UP000078540 UP000005205 UP000242457 UP000005203 UP000015103 UP000095301 UP000078542 UP000000304 UP000008237 UP000192221 UP000000803 UP000002282 UP000009046 UP000008711 UP000008792 UP000001292 UP000007798 UP000095300 UP000000311 UP000000673 UP000027135 UP000001070 UP000092553 UP000075880 UP000075920 UP000075886 UP000007755 UP000008820 UP000076407 UP000075903 UP000075840 UP000007062 UP000000305 UP000053825 UP000268350 UP000008744 UP000075883 UP000015104 UP000019118 UP000030742 UP000001819 UP000076858 UP000002320 UP000009192 UP000069940 UP000249989 UP000285301 UP000030765 UP000076408 UP000075884 UP000183832 UP000075900 UP000007266 UP000094527 UP000075885
UP000235965 UP000053097 UP000279307 UP000036403 UP000192223 UP000078541 UP000076502 UP000078492 UP000037069 UP000075809 UP000007801 UP000078540 UP000005205 UP000242457 UP000005203 UP000015103 UP000095301 UP000078542 UP000000304 UP000008237 UP000192221 UP000000803 UP000002282 UP000009046 UP000008711 UP000008792 UP000001292 UP000007798 UP000095300 UP000000311 UP000000673 UP000027135 UP000001070 UP000092553 UP000075880 UP000075920 UP000075886 UP000007755 UP000008820 UP000076407 UP000075903 UP000075840 UP000007062 UP000000305 UP000053825 UP000268350 UP000008744 UP000075883 UP000015104 UP000019118 UP000030742 UP000001819 UP000076858 UP000002320 UP000009192 UP000069940 UP000249989 UP000285301 UP000030765 UP000076408 UP000075884 UP000183832 UP000075900 UP000007266 UP000094527 UP000075885
PRIDE
Interpro
Gene 3D
ProteinModelPortal
H9JJI5
A0A0L7KP52
A0A1E1W0G5
A0A2A4JGG7
A0A2H1WS05
A0A0N0PEF8
+ More
A0A194Q5G6 A0A212EMI0 A0A2W1BIL1 A0A2J7QMU3 A0A026WIJ9 A0A0J7NP49 E9IA29 A0A1W4X8D1 A0A0T6ATU0 A0A195FKW4 A0A154P0V2 A0A151J2Z6 A0A0L0BXI9 A0A151X0K3 B3MBH6 A0A0K8TLW6 A0A151I3S8 A0A158NVF3 A0A2A3ELG8 A0A088A139 T1HNB3 T1PI23 A0A1Y1KC89 U5ET80 A0A151I660 B4QH16 E2BSI3 A0A0C9QWI4 A0A1W4UN08 Q9W297 B4P8B1 A0A0P4VSD5 E0VZ07 B3NNC4 A0A0A9VVY6 B4LNE0 B4I7Z5 B4MP51 A0A1I8NWI7 A0A2M4AQM3 E2AW08 W5JFU9 A0A336LLW1 A0A0K8SVR4 A0A067QL28 A0A2R7WAX0 B4J8K0 A0A1L8DYS8 A0A069DSP4 A0A0M4E8T4 A0A224XE19 A0A182IVY9 A0A1Y9IWB2 A0A0V0GC91 A0A1B6D9E7 A0A182QAP4 A0A0A1XFH9 A0A034WP16 F4WR79 A0A1B6J1T7 Q17NM2 A0A3F2Z244 A0A2Y9D2X6 A0A2C9GQZ1 A0NGK2 E9HDH0 A0A0L7QU93 A0A3B0JWC1 B4GHB2 A0A182LWA6 T1KRP3 T1DI74 N6T957 A0A0P5ZIB4 Q28XN6 A0A0N8ECY0 B0XJR8 A0A0P6BUY5 B4KMV8 A0A1Q3F784 A0A1Q3F792 A0A182GNX7 A0A3S3QD11 A0A084WIW5 A0A182Y1G3 A0A182MYU6 W8C0T2 T1JHN0 A0A1J1JA77 A0A182RD16 D7EIL2 A0A1D2N9A2 A0A182P5V0 A0A0P6I429
A0A194Q5G6 A0A212EMI0 A0A2W1BIL1 A0A2J7QMU3 A0A026WIJ9 A0A0J7NP49 E9IA29 A0A1W4X8D1 A0A0T6ATU0 A0A195FKW4 A0A154P0V2 A0A151J2Z6 A0A0L0BXI9 A0A151X0K3 B3MBH6 A0A0K8TLW6 A0A151I3S8 A0A158NVF3 A0A2A3ELG8 A0A088A139 T1HNB3 T1PI23 A0A1Y1KC89 U5ET80 A0A151I660 B4QH16 E2BSI3 A0A0C9QWI4 A0A1W4UN08 Q9W297 B4P8B1 A0A0P4VSD5 E0VZ07 B3NNC4 A0A0A9VVY6 B4LNE0 B4I7Z5 B4MP51 A0A1I8NWI7 A0A2M4AQM3 E2AW08 W5JFU9 A0A336LLW1 A0A0K8SVR4 A0A067QL28 A0A2R7WAX0 B4J8K0 A0A1L8DYS8 A0A069DSP4 A0A0M4E8T4 A0A224XE19 A0A182IVY9 A0A1Y9IWB2 A0A0V0GC91 A0A1B6D9E7 A0A182QAP4 A0A0A1XFH9 A0A034WP16 F4WR79 A0A1B6J1T7 Q17NM2 A0A3F2Z244 A0A2Y9D2X6 A0A2C9GQZ1 A0NGK2 E9HDH0 A0A0L7QU93 A0A3B0JWC1 B4GHB2 A0A182LWA6 T1KRP3 T1DI74 N6T957 A0A0P5ZIB4 Q28XN6 A0A0N8ECY0 B0XJR8 A0A0P6BUY5 B4KMV8 A0A1Q3F784 A0A1Q3F792 A0A182GNX7 A0A3S3QD11 A0A084WIW5 A0A182Y1G3 A0A182MYU6 W8C0T2 T1JHN0 A0A1J1JA77 A0A182RD16 D7EIL2 A0A1D2N9A2 A0A182P5V0 A0A0P6I429
PDB
5EJZ
E-value=0.0530474,
Score=85
Ontologies
KEGG
PATHWAY
GO
PANTHER
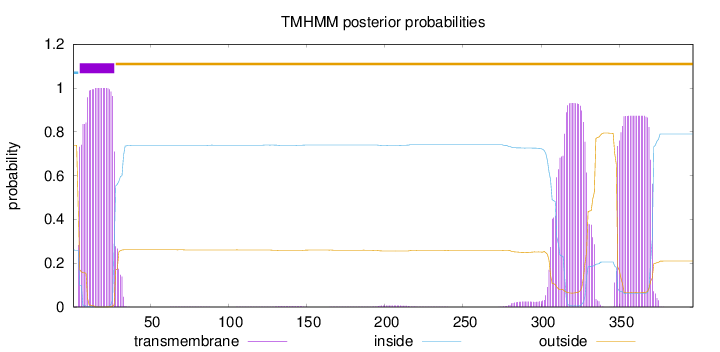
Topology
Length:
397
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
63.3827200000001
Exp number, first 60 AAs:
22.90353
Total prob of N-in:
0.26179
POSSIBLE N-term signal
sequence
inside
1 - 4
TMhelix
5 - 27
outside
28 - 397
Population Genetic Test Statistics
Pi
75.19028
Theta
153.454516
Tajima's D
-1.601831
CLR
18.435381
CSRT
0.0491475426228689
Interpretation
Uncertain
