Gene
KWMTBOMO12106
Pre Gene Modal
BGIBMGA004353
Annotation
PREDICTED:_RNA-binding_protein_fusilli_isoform_X3_[Papilio_xuthus]
Location in the cell
Mitochondrial Reliability : 1.664 Nuclear Reliability : 1.195
Sequence
CDS
ATGGCTCTGAAGCGGCATAAACACCACATCGGCCCTCGGTACATTGAGGTGTACCGGGCGAGTGGTGAAGATTTCCTCTCGGTGGCTGGTGGCGCGACCTGTGAAGCAGCTGCCTTCCTATCGCGAGGAGCGCGCGTCATCGTCAGAATGCGAGGATTACCTTACGATGCCACTGCTCAACAAGTCTTGGATTTCTTCACGAGCGGCGAGGACCCAGTCCAGGTCCTGGATGGTACTGAGGGAGTGCTGTTTGTGCGTCGCGCCGACGGTCGCGCGACCGGCGACGCTTTCGTGCTTTTCGCGAAAGAGGAGGACTCACCCAAAGCCCTGTCGCGACATCGCAAGCTCATCGGGTCTCGATACATCGAACTCTTTCGCTCCACCATCGCTGAAGTGCAGCAGGTCCTAAACCGCTCGTTGGAGAGTCGTACAAGCCAAACCAGCACCCCGTCCACGGCCCCCGCTCCCGCTGAAGGTCTTCTGCCAGTGGCGTTGGTGCCACAGCACGTTATCACATCTGGAACCGCCAAAGACTGCGTTAGACTGCGTGGACTGCCTTATGAAGCCCAGGTTGAACACATCCTGACGTTTCTGGAAGAATTCGCCAAGAACATTGTCGTGCAAGGAGTTCACATGGTTTTCAATGCCCAGGGTCATCCCAGCGGCGAGGCATTCATTCAGATGGACAGCGAGGCCAGCGCTTTCCTCTGTGCCCAGCAGAAGCATCACCGGTACATGACCTTCGGGAAGAAACAACGGTACATCGAGGTGTTCCAGTGTTCAGGCGACGACATGAACATGGTGCTAACGGGCGGCGTGACCCCGGCGACGTCCCCGAAGGTGTTGTCCCCCGGTCCTTTGACTTATTACTATTCAGCAATGGGACCGACGCTGCCACCCCCGCCGCTCGTATATTGGGGCTACCCGACACCCCCAGTCTCCCCTGCTCACTATTACCACCCTCCACACCATCCGCAAACCATGGTTAATTTTATACAAATAAGGTTAATTTTATACAAATAA
Protein
MALKRHKHHIGPRYIEVYRASGEDFLSVAGGATCEAAAFLSRGARVIVRMRGLPYDATAQQVLDFFTSGEDPVQVLDGTEGVLFVRRADGRATGDAFVLFAKEEDSPKALSRHRKLIGSRYIELFRSTIAEVQQVLNRSLESRTSQTSTPSTAPAPAEGLLPVALVPQHVITSGTAKDCVRLRGLPYEAQVEHILTFLEEFAKNIVVQGVHMVFNAQGHPSGEAFIQMDSEASAFLCAQQKHHRYMTFGKKQRYIEVFQCSGDDMNMVLTGGVTPATSPKVLSPGPLTYYYSAMGPTLPPPPLVYWGYPTPPVSPAHYYHPPHHPQTMVNFIQIRLILYK
Summary
Uniprot
A0A3S2L409
S4NVL4
A0A2A4JMR5
A0A2W1CP21
A0A2H1W5I1
A0A194Q5H7
+ More
A0A1E1W1X5 A0A0N1IQ32 H9J4B4 A0A2A4JNW7 A0A1Y1MBC0 A0A0T6B5P2 A0A1Y1MAP4 V5I8K1 A0A1L8DGE4 A0A2J7R0Z2 V5GQR4 A0A2J7R0X6 A0A2J7R0Y6 A0A1L8DGR4 D6WSB8 A0A139WFL7 A0A0A1WY29 A0A2J7R0Y5 A0A2J7R0Z1 A0A2J7R0Y3 E2ABC3 A0A2J7R0Y9 V9IBQ8 E9JD32 A0A1B0A7D3 A0A158NWT2 A0A0K8UJ53 A0A0J7KU84 A0A1A9VXG7 A0A0K8W3G7 W8AYV1 A0A034WBC5 A0A2P8XTD5 A0A1B6EGH5 A0A1B6D759 A0A2A3EPD3 A0A034W8Q1 A0A151JPJ4 A0A1A9Y433 A0A1B0BI53 F4W9C6 A0A195FX22 A0A2J7R0Z0 A0A0J9U2V6 A0A0A1WF22 A0A0P4W8T0 A0A1B6I734 A0A1B6MRX3 A0A154PS49 A0A1B0GAA9 A0A0P4VWB4 A0A0L7RH50 A0A1A9W9T1 A0A0M8ZPD7 A0A0A9YIV9 A0A1B6CA42 A0A2M4A7L4 A0A2M4A7D3 A0A0B4K880 A0A1B6LBY8 A0A0K8VW31 A0A1I8NAZ1 A0A2M4BD11 A0A0R3NVS5 A0A146LIN5 A0A0A9YHB2 A0A026WNJ7 A0A161MZW7 A0A1Q3G348 A0A224XN77 A0A1Q3G335 A0A0P4W3N9 A0A0K8VMH0 A0A1I8NB03 A0A3L8DVT1 A0A182VNN1 A0A1I8P1A6 W8ANQ0 A0A0A9YPU4 A0A0A9YKC6 A0A0P4W3N2 A0A067RKA5 A0A182H406 A0A0C9RKR2 A0A3B0IYA1 A0A2M4CXC5 A0A182FU22 A0A0J9RDI3 B0WRI7 A0A0Q9XHJ8 A0A069DWC1 W5JJP3 A0A1W4WK73 A0A2M4CYI3
A0A1E1W1X5 A0A0N1IQ32 H9J4B4 A0A2A4JNW7 A0A1Y1MBC0 A0A0T6B5P2 A0A1Y1MAP4 V5I8K1 A0A1L8DGE4 A0A2J7R0Z2 V5GQR4 A0A2J7R0X6 A0A2J7R0Y6 A0A1L8DGR4 D6WSB8 A0A139WFL7 A0A0A1WY29 A0A2J7R0Y5 A0A2J7R0Z1 A0A2J7R0Y3 E2ABC3 A0A2J7R0Y9 V9IBQ8 E9JD32 A0A1B0A7D3 A0A158NWT2 A0A0K8UJ53 A0A0J7KU84 A0A1A9VXG7 A0A0K8W3G7 W8AYV1 A0A034WBC5 A0A2P8XTD5 A0A1B6EGH5 A0A1B6D759 A0A2A3EPD3 A0A034W8Q1 A0A151JPJ4 A0A1A9Y433 A0A1B0BI53 F4W9C6 A0A195FX22 A0A2J7R0Z0 A0A0J9U2V6 A0A0A1WF22 A0A0P4W8T0 A0A1B6I734 A0A1B6MRX3 A0A154PS49 A0A1B0GAA9 A0A0P4VWB4 A0A0L7RH50 A0A1A9W9T1 A0A0M8ZPD7 A0A0A9YIV9 A0A1B6CA42 A0A2M4A7L4 A0A2M4A7D3 A0A0B4K880 A0A1B6LBY8 A0A0K8VW31 A0A1I8NAZ1 A0A2M4BD11 A0A0R3NVS5 A0A146LIN5 A0A0A9YHB2 A0A026WNJ7 A0A161MZW7 A0A1Q3G348 A0A224XN77 A0A1Q3G335 A0A0P4W3N9 A0A0K8VMH0 A0A1I8NB03 A0A3L8DVT1 A0A182VNN1 A0A1I8P1A6 W8ANQ0 A0A0A9YPU4 A0A0A9YKC6 A0A0P4W3N2 A0A067RKA5 A0A182H406 A0A0C9RKR2 A0A3B0IYA1 A0A2M4CXC5 A0A182FU22 A0A0J9RDI3 B0WRI7 A0A0Q9XHJ8 A0A069DWC1 W5JJP3 A0A1W4WK73 A0A2M4CYI3
Pubmed
23622113
28756777
26354079
19121390
28004739
18362917
+ More
19820115 25830018 20798317 21282665 21347285 24495485 25348373 29403074 21719571 22936249 25401762 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 25315136 15632085 17994087 26823975 24508170 30249741 24845553 26483478 26334808 20920257 23761445
19820115 25830018 20798317 21282665 21347285 24495485 25348373 29403074 21719571 22936249 25401762 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 25315136 15632085 17994087 26823975 24508170 30249741 24845553 26483478 26334808 20920257 23761445
EMBL
RSAL01000172
RVE45104.1
GAIX01011491
JAA81069.1
NWSH01001014
PCG73109.1
+ More
NHMN01023844 PZC87385.1 ODYU01006157 SOQ47764.1 KQ459460 KPJ00798.1 GDQN01010056 JAT80998.1 KQ459838 KPJ19722.1 BABH01041341 PCG73110.1 GEZM01036199 GEZM01036198 JAV82901.1 LJIG01009637 KRT82681.1 GEZM01036194 JAV82909.1 GALX01004545 JAB63921.1 GFDF01008568 JAV05516.1 NEVH01008216 PNF34495.1 GALX01004544 JAB63922.1 PNF34490.1 PNF34491.1 GFDF01008438 JAV05646.1 KQ971352 EFA06390.1 KYB26597.1 GBXI01010716 JAD03576.1 PNF34498.1 PNF34496.1 PNF34494.1 GL438237 EFN69244.1 PNF34492.1 JR037395 AEY57749.1 GL771866 EFZ09344.1 ADTU01028468 GDHF01025736 JAI26578.1 LBMM01003158 KMQ93883.1 GDHF01014259 GDHF01006685 JAI38055.1 JAI45629.1 GAMC01020464 JAB86091.1 GAKP01007869 GAKP01007866 JAC51083.1 PYGN01001378 PSN35277.1 GEDC01000252 JAS37046.1 GEDC01015777 JAS21521.1 KZ288206 PBC33066.1 GAKP01007868 GAKP01007867 GAKP01007865 JAC51084.1 KQ978718 KYN29054.1 JXJN01014772 JXJN01014773 GL888002 EGI69308.1 KQ981204 KYN44976.1 PNF34497.1 CM002911 KMY94040.1 GBXI01016795 JAC97496.1 GDRN01091681 GDRN01091675 JAI60238.1 GECU01025006 JAS82700.1 GEBQ01001280 JAT38697.1 KQ435037 KZC14088.1 CCAG010006819 CCAG010006820 GDRN01091686 JAI60235.1 KQ414596 KOC70031.1 KQ435944 KOX68207.1 GBHO01012053 GBRD01014752 GBRD01014751 JAG31551.1 JAG51074.1 GEDC01026955 GEDC01013842 JAS10343.1 JAS23456.1 GGFK01003399 MBW36720.1 GGFK01003395 MBW36716.1 AE013599 AFH08101.1 GEBQ01018774 JAT21203.1 GDHF01009217 JAI43097.1 GGFJ01001791 MBW50932.1 CM000071 KRT03293.1 GDHC01010635 JAQ07994.1 GBHO01012055 JAG31549.1 KK107151 EZA57241.1 GEMB01003147 JAS00062.1 GFDL01000822 JAV34223.1 GFTR01006536 JAW09890.1 GFDL01000832 JAV34213.1 GDRN01091678 GDRN01091677 JAI60242.1 GDHF01012594 JAI39720.1 QOIP01000003 RLU24531.1 GAMC01020467 GAMC01020466 GAMC01020465 GAMC01020463 JAB86088.1 GBHO01012054 JAG31550.1 GBHO01012051 GBRD01015566 GBRD01014754 GBRD01014750 GBRD01011529 JAG31553.1 JAG50260.1 GDRN01091689 JAI60233.1 KK852423 KDR24242.1 JXUM01108447 JXUM01108448 KQ565231 KXJ71204.1 GBYB01013882 GBYB01013883 JAG83649.1 JAG83650.1 OUUW01000001 SPP72707.1 GGFL01005741 MBW69919.1 KMY94042.1 DS232056 EDS33385.1 CH933808 KRG05354.1 GBGD01000748 JAC88141.1 ADMH02001328 ETN62989.1 GGFL01005740 MBW69918.1
NHMN01023844 PZC87385.1 ODYU01006157 SOQ47764.1 KQ459460 KPJ00798.1 GDQN01010056 JAT80998.1 KQ459838 KPJ19722.1 BABH01041341 PCG73110.1 GEZM01036199 GEZM01036198 JAV82901.1 LJIG01009637 KRT82681.1 GEZM01036194 JAV82909.1 GALX01004545 JAB63921.1 GFDF01008568 JAV05516.1 NEVH01008216 PNF34495.1 GALX01004544 JAB63922.1 PNF34490.1 PNF34491.1 GFDF01008438 JAV05646.1 KQ971352 EFA06390.1 KYB26597.1 GBXI01010716 JAD03576.1 PNF34498.1 PNF34496.1 PNF34494.1 GL438237 EFN69244.1 PNF34492.1 JR037395 AEY57749.1 GL771866 EFZ09344.1 ADTU01028468 GDHF01025736 JAI26578.1 LBMM01003158 KMQ93883.1 GDHF01014259 GDHF01006685 JAI38055.1 JAI45629.1 GAMC01020464 JAB86091.1 GAKP01007869 GAKP01007866 JAC51083.1 PYGN01001378 PSN35277.1 GEDC01000252 JAS37046.1 GEDC01015777 JAS21521.1 KZ288206 PBC33066.1 GAKP01007868 GAKP01007867 GAKP01007865 JAC51084.1 KQ978718 KYN29054.1 JXJN01014772 JXJN01014773 GL888002 EGI69308.1 KQ981204 KYN44976.1 PNF34497.1 CM002911 KMY94040.1 GBXI01016795 JAC97496.1 GDRN01091681 GDRN01091675 JAI60238.1 GECU01025006 JAS82700.1 GEBQ01001280 JAT38697.1 KQ435037 KZC14088.1 CCAG010006819 CCAG010006820 GDRN01091686 JAI60235.1 KQ414596 KOC70031.1 KQ435944 KOX68207.1 GBHO01012053 GBRD01014752 GBRD01014751 JAG31551.1 JAG51074.1 GEDC01026955 GEDC01013842 JAS10343.1 JAS23456.1 GGFK01003399 MBW36720.1 GGFK01003395 MBW36716.1 AE013599 AFH08101.1 GEBQ01018774 JAT21203.1 GDHF01009217 JAI43097.1 GGFJ01001791 MBW50932.1 CM000071 KRT03293.1 GDHC01010635 JAQ07994.1 GBHO01012055 JAG31549.1 KK107151 EZA57241.1 GEMB01003147 JAS00062.1 GFDL01000822 JAV34223.1 GFTR01006536 JAW09890.1 GFDL01000832 JAV34213.1 GDRN01091678 GDRN01091677 JAI60242.1 GDHF01012594 JAI39720.1 QOIP01000003 RLU24531.1 GAMC01020467 GAMC01020466 GAMC01020465 GAMC01020463 JAB86088.1 GBHO01012054 JAG31550.1 GBHO01012051 GBRD01015566 GBRD01014754 GBRD01014750 GBRD01011529 JAG31553.1 JAG50260.1 GDRN01091689 JAI60233.1 KK852423 KDR24242.1 JXUM01108447 JXUM01108448 KQ565231 KXJ71204.1 GBYB01013882 GBYB01013883 JAG83649.1 JAG83650.1 OUUW01000001 SPP72707.1 GGFL01005741 MBW69919.1 KMY94042.1 DS232056 EDS33385.1 CH933808 KRG05354.1 GBGD01000748 JAC88141.1 ADMH02001328 ETN62989.1 GGFL01005740 MBW69918.1
Proteomes
UP000283053
UP000218220
UP000053268
UP000053240
UP000005204
UP000235965
+ More
UP000007266 UP000000311 UP000092445 UP000005205 UP000036403 UP000078200 UP000245037 UP000242457 UP000078492 UP000092443 UP000092460 UP000007755 UP000078541 UP000076502 UP000092444 UP000053825 UP000091820 UP000053105 UP000000803 UP000095301 UP000001819 UP000053097 UP000279307 UP000075903 UP000095300 UP000027135 UP000069940 UP000249989 UP000268350 UP000069272 UP000002320 UP000009192 UP000000673 UP000192223
UP000007266 UP000000311 UP000092445 UP000005205 UP000036403 UP000078200 UP000245037 UP000242457 UP000078492 UP000092443 UP000092460 UP000007755 UP000078541 UP000076502 UP000092444 UP000053825 UP000091820 UP000053105 UP000000803 UP000095301 UP000001819 UP000053097 UP000279307 UP000075903 UP000095300 UP000027135 UP000069940 UP000249989 UP000268350 UP000069272 UP000002320 UP000009192 UP000000673 UP000192223
PRIDE
Interpro
Gene 3D
ProteinModelPortal
A0A3S2L409
S4NVL4
A0A2A4JMR5
A0A2W1CP21
A0A2H1W5I1
A0A194Q5H7
+ More
A0A1E1W1X5 A0A0N1IQ32 H9J4B4 A0A2A4JNW7 A0A1Y1MBC0 A0A0T6B5P2 A0A1Y1MAP4 V5I8K1 A0A1L8DGE4 A0A2J7R0Z2 V5GQR4 A0A2J7R0X6 A0A2J7R0Y6 A0A1L8DGR4 D6WSB8 A0A139WFL7 A0A0A1WY29 A0A2J7R0Y5 A0A2J7R0Z1 A0A2J7R0Y3 E2ABC3 A0A2J7R0Y9 V9IBQ8 E9JD32 A0A1B0A7D3 A0A158NWT2 A0A0K8UJ53 A0A0J7KU84 A0A1A9VXG7 A0A0K8W3G7 W8AYV1 A0A034WBC5 A0A2P8XTD5 A0A1B6EGH5 A0A1B6D759 A0A2A3EPD3 A0A034W8Q1 A0A151JPJ4 A0A1A9Y433 A0A1B0BI53 F4W9C6 A0A195FX22 A0A2J7R0Z0 A0A0J9U2V6 A0A0A1WF22 A0A0P4W8T0 A0A1B6I734 A0A1B6MRX3 A0A154PS49 A0A1B0GAA9 A0A0P4VWB4 A0A0L7RH50 A0A1A9W9T1 A0A0M8ZPD7 A0A0A9YIV9 A0A1B6CA42 A0A2M4A7L4 A0A2M4A7D3 A0A0B4K880 A0A1B6LBY8 A0A0K8VW31 A0A1I8NAZ1 A0A2M4BD11 A0A0R3NVS5 A0A146LIN5 A0A0A9YHB2 A0A026WNJ7 A0A161MZW7 A0A1Q3G348 A0A224XN77 A0A1Q3G335 A0A0P4W3N9 A0A0K8VMH0 A0A1I8NB03 A0A3L8DVT1 A0A182VNN1 A0A1I8P1A6 W8ANQ0 A0A0A9YPU4 A0A0A9YKC6 A0A0P4W3N2 A0A067RKA5 A0A182H406 A0A0C9RKR2 A0A3B0IYA1 A0A2M4CXC5 A0A182FU22 A0A0J9RDI3 B0WRI7 A0A0Q9XHJ8 A0A069DWC1 W5JJP3 A0A1W4WK73 A0A2M4CYI3
A0A1E1W1X5 A0A0N1IQ32 H9J4B4 A0A2A4JNW7 A0A1Y1MBC0 A0A0T6B5P2 A0A1Y1MAP4 V5I8K1 A0A1L8DGE4 A0A2J7R0Z2 V5GQR4 A0A2J7R0X6 A0A2J7R0Y6 A0A1L8DGR4 D6WSB8 A0A139WFL7 A0A0A1WY29 A0A2J7R0Y5 A0A2J7R0Z1 A0A2J7R0Y3 E2ABC3 A0A2J7R0Y9 V9IBQ8 E9JD32 A0A1B0A7D3 A0A158NWT2 A0A0K8UJ53 A0A0J7KU84 A0A1A9VXG7 A0A0K8W3G7 W8AYV1 A0A034WBC5 A0A2P8XTD5 A0A1B6EGH5 A0A1B6D759 A0A2A3EPD3 A0A034W8Q1 A0A151JPJ4 A0A1A9Y433 A0A1B0BI53 F4W9C6 A0A195FX22 A0A2J7R0Z0 A0A0J9U2V6 A0A0A1WF22 A0A0P4W8T0 A0A1B6I734 A0A1B6MRX3 A0A154PS49 A0A1B0GAA9 A0A0P4VWB4 A0A0L7RH50 A0A1A9W9T1 A0A0M8ZPD7 A0A0A9YIV9 A0A1B6CA42 A0A2M4A7L4 A0A2M4A7D3 A0A0B4K880 A0A1B6LBY8 A0A0K8VW31 A0A1I8NAZ1 A0A2M4BD11 A0A0R3NVS5 A0A146LIN5 A0A0A9YHB2 A0A026WNJ7 A0A161MZW7 A0A1Q3G348 A0A224XN77 A0A1Q3G335 A0A0P4W3N9 A0A0K8VMH0 A0A1I8NB03 A0A3L8DVT1 A0A182VNN1 A0A1I8P1A6 W8ANQ0 A0A0A9YPU4 A0A0A9YKC6 A0A0P4W3N2 A0A067RKA5 A0A182H406 A0A0C9RKR2 A0A3B0IYA1 A0A2M4CXC5 A0A182FU22 A0A0J9RDI3 B0WRI7 A0A0Q9XHJ8 A0A069DWC1 W5JJP3 A0A1W4WK73 A0A2M4CYI3
PDB
2RVJ
E-value=1.69078e-27,
Score=304
Ontologies
KEGG
GO
PANTHER
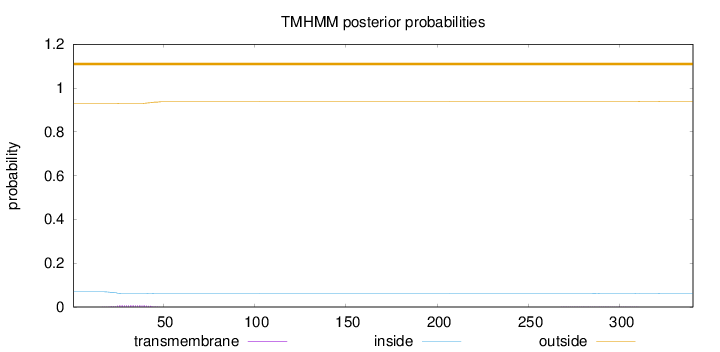
Topology
Length:
340
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.18777
Exp number, first 60 AAs:
0.16181
Total prob of N-in:
0.06929
outside
1 - 340
Population Genetic Test Statistics
Pi
185.276304
Theta
168.8314
Tajima's D
-1.429081
CLR
1328.563562
CSRT
0.0656967151642418
Interpretation
Uncertain
