Gene
KWMTBOMO12102
Pre Gene Modal
BGIBMGA004270
Annotation
origin_recognition_complex_subunit_6_[Bombyx_mori]
Full name
Origin recognition complex subunit 6
Location in the cell
Nuclear Reliability : 3.084
Sequence
CDS
ATGGCGAAGTACAATAAAACTTTACATCTCATAGCTTCCAAACTTGGTATCAGTGACGAAGAAAAAGTTTTAAGGAAAGCAGCTGAGTTTGAACGACTCTTAGAAACAAAATCTACGGCTGGCAGCAACATTACTGATACAAGTAAAGTTGTTATATGCTTGGACTTGGCTGCTAGTTTGTATGGTACTGAATTTGATGTTAAGCAAGCAATTAAGTATTCTGGTCTGAGGCCAACAGTGTATGCAAACAGCAAGAAAATAATATCAAATCGACTAGAATTGGATGTCGATAGACTCAGTGTTGCAGTTCTTTGTGCTAGCTTACAAAACAGTGGTGTTCAGGCGCTAGCTGAGAAAATACTTGAGCAGTACAAGCAGTACAGTAAAGTAGAATTGGATATGAGTCTTCCTCAGTATGTGTGTATGGCTGTTTACCAAGCTTGCAGGATAAGCAAAGTTAAAGTATCAAAGAGTAAAATAATTGAAAAGGCTAGATTAAAGCCAGCCCAGTGGAATAAATTAGAAGCTGACTGGAGTCAATTTGCCGATACACACTTTGCGGTCAAAAAGAGAGGCAAGAAAGAAGTTAAAAATTCTATTGAAAACGTCGAAGAAAATGTACCAATGGAAGTGGACAGTAGTAAACAAGAGGAGCCAATTGAAACATTTGAACCGTACGAACAGTGGAAGAAACGAATGCTCGATTCTGCGTATTCAGAGTTAAAGGAGCTAGAGAAATCACAAAGCGACAAGTTGAAGGATATGGTGATGTCACCAAGAAGGTCTCCACGTAAAACACCACAAAAGTATTCTCCGTATAAAAGTTTGAATACAAAAGGTAGTGTGCGGCTGCTTTTTCCCATTGAAAGTAACTAA
Protein
MAKYNKTLHLIASKLGISDEEKVLRKAAEFERLLETKSTAGSNITDTSKVVICLDLAASLYGTEFDVKQAIKYSGLRPTVYANSKKIISNRLELDVDRLSVAVLCASLQNSGVQALAEKILEQYKQYSKVELDMSLPQYVCMAVYQACRISKVKVSKSKIIEKARLKPAQWNKLEADWSQFADTHFAVKKRGKKEVKNSIENVEENVPMEVDSSKQEEPIETFEPYEQWKKRMLDSAYSELKELEKSQSDKLKDMVMSPRRSPRKTPQKYSPYKSLNTKGSVRLLFPIESN
Summary
Description
Component of the origin recognition complex (ORC) that binds origins of replication. DNA-binding is ATP-dependent, however specific DNA sequences that define origins of replication have not been identified so far. ORC is required to assemble the pre-replication complex necessary to initiate DNA replication (By similarity).
Component of the origin recognition complex (ORC) that binds origins of replication. DNA-binding is ATP-dependent. The specific DNA sequences that define origins of replication have not been identified yet. ORC is required to assemble the pre-replication complex necessary to initiate DNA replication (By similarity).
Component of the origin recognition complex (ORC) that binds origins of replication. DNA-binding is ATP-dependent. The specific DNA sequences that define origins of replication have not been identified yet. ORC is required to assemble the pre-replication complex necessary to initiate DNA replication (By similarity).
Subunit
ORC is composed of six subunits.
Component of ORC, a complex composed of at least 6 subunits: ORC1, ORC2, ORC3, ORC4, ORC5 and ORC6. ORC is regulated in a cell-cycle dependent manner. It is sequentially assembled at the exit from anaphase of mitosis and disassembled as cells enter S phase (By similarity). Interacts with DBF4 (By similarity).
Component of ORC, a complex composed of at least 6 subunits: ORC1, ORC2, ORC3, ORC4, ORC5 and ORC6. ORC is regulated in a cell-cycle dependent manner. It is sequentially assembled at the exit from anaphase of mitosis and disassembled as cells enter S phase (By similarity). Interacts with DBF4 (By similarity).
Similarity
Belongs to the ORC6 family.
Keywords
3D-structure
Complete proteome
DNA replication
DNA-binding
Nucleus
Reference proteome
Isopeptide bond
Phosphoprotein
Ubl conjugation
Feature
chain Origin recognition complex subunit 6
Uniprot
H9J431
D3VW61
A0A2A4K691
A0A2W1BLJ8
A0A0N1PIA9
S4NIE2
+ More
A6N831 A0A2H1X1P7 A0A212ELK0 A0A194Q728 A0A182K810 A0A182M6L8 A0A182Q437 A0A084VEW6 A0A182WDB5 A0A182IK57 A0A182P353 B0WD73 W5J968 A0A182YE87 A0A1Q3EVG5 Q16TG1 A0A1S4FQ03 Q7PSQ8 A0A182WR78 A0A182VFA2 A0A182MXF9 A0A182RXM7 A0A182HEY2 A0A182FUW9 A0A182HG20 A0A182TY94 B4MKB2 A0A1I8M4H2 B3N6V2 A0A182LHI8 A0A0J9R928 Q9Y1B2 A0A1W4UZ81 B4NWL7 B3MGP7 B4QHS0 A0A1I8P103 B4INL9 A0A1B0FMY7 A0A1A9ZD43 B4KMG7 A0A1A9UYX7 A0A1A9XLI1 A0A1B0BAQ1 A0A1A9W741 A0A1J1IT23 B4LNG3 A0A0M5IXY8 A0A336LP92 A0A3B0JHX9 A0A1B0GJG5 B4H588 B5E187 B4J8A4 A0A1W4XKD0 A0A3R7N652 A0A210QH20 A0A0K8WLN3 A0A034V5D3 U4UHE6 B4K2P2 N6UFC8 A0A2P8XL04 A0A1B6FT33 A0A2J7QJQ8 A0A093GZB4 B5G1W7 A0A1B6LPB1 A0A3L5TTG3 A0A383YXV5 A0A218UWG5 A0A3L8RZK0 H0ZEY5 I3LLV3 A0A341BYV8 A0A1S3ARQ1 A0A1U7SEN7 L8IRR5 Q2HJF3 B5G1W6 A0A2Y9DBW2 E9GM69 A0A2K6EW96 A0A091P2T6 F7EFU1 F7E7B7
A6N831 A0A2H1X1P7 A0A212ELK0 A0A194Q728 A0A182K810 A0A182M6L8 A0A182Q437 A0A084VEW6 A0A182WDB5 A0A182IK57 A0A182P353 B0WD73 W5J968 A0A182YE87 A0A1Q3EVG5 Q16TG1 A0A1S4FQ03 Q7PSQ8 A0A182WR78 A0A182VFA2 A0A182MXF9 A0A182RXM7 A0A182HEY2 A0A182FUW9 A0A182HG20 A0A182TY94 B4MKB2 A0A1I8M4H2 B3N6V2 A0A182LHI8 A0A0J9R928 Q9Y1B2 A0A1W4UZ81 B4NWL7 B3MGP7 B4QHS0 A0A1I8P103 B4INL9 A0A1B0FMY7 A0A1A9ZD43 B4KMG7 A0A1A9UYX7 A0A1A9XLI1 A0A1B0BAQ1 A0A1A9W741 A0A1J1IT23 B4LNG3 A0A0M5IXY8 A0A336LP92 A0A3B0JHX9 A0A1B0GJG5 B4H588 B5E187 B4J8A4 A0A1W4XKD0 A0A3R7N652 A0A210QH20 A0A0K8WLN3 A0A034V5D3 U4UHE6 B4K2P2 N6UFC8 A0A2P8XL04 A0A1B6FT33 A0A2J7QJQ8 A0A093GZB4 B5G1W7 A0A1B6LPB1 A0A3L5TTG3 A0A383YXV5 A0A218UWG5 A0A3L8RZK0 H0ZEY5 I3LLV3 A0A341BYV8 A0A1S3ARQ1 A0A1U7SEN7 L8IRR5 Q2HJF3 B5G1W6 A0A2Y9DBW2 E9GM69 A0A2K6EW96 A0A091P2T6 F7EFU1 F7E7B7
Pubmed
19121390
21162711
28756777
26354079
23622113
17129416
+ More
22118469 24438588 20920257 23761445 25244985 17510324 12364791 26483478 17994087 25315136 20966253 22936249 10346817 10731132 12537572 12537569 17550304 15632085 28812685 25348373 23537049 29403074 17018643 26977809 30282656 20360741 22751099 21292972 17495919 18464734
22118469 24438588 20920257 23761445 25244985 17510324 12364791 26483478 17994087 25315136 20966253 22936249 10346817 10731132 12537572 12537569 17550304 15632085 28812685 25348373 23537049 29403074 17018643 26977809 30282656 20360741 22751099 21292972 17495919 18464734
EMBL
BABH01032651
GQ214395
ADD10142.1
NWSH01000127
PCG79283.1
KZ150043
+ More
PZC74535.1 KQ459838 KPJ19717.1 GAIX01014109 JAA78451.1 EF619926 ABR09538.1 ODYU01012760 SOQ59229.1 AGBW02014044 OWR42353.1 KQ459460 KPJ00805.1 AXCM01007145 AXCN02001700 ATLV01012340 KE524783 KFB36510.1 DS231894 EDS44251.1 ADMH02001912 ETN60531.1 GFDL01015752 JAV19293.1 CH477650 EAT37789.1 AAAB01008816 EAA05187.5 JXUM01133026 KQ567923 KXJ69230.1 APCN01005461 CH963846 EDW72551.1 CH954177 EDV58201.1 CM002911 KMY92588.1 AF139064 AE013599 AY071443 CM000157 EDW89562.2 CH902619 EDV36805.1 CM000362 EDX06404.1 CH676472 EDW49402.1 CCAG010000346 CH933808 EDW09855.1 JXJN01011105 CVRI01000059 CRL03359.1 CH940648 EDW61115.1 CP012524 ALC42424.1 UFQT01000040 SSX18611.1 OUUW01000001 SPP74990.1 AJWK01020294 CH479210 EDW32924.1 CM000071 EDY69475.1 CH916367 EDW01241.1 QCYY01001403 ROT78273.1 NEDP02003738 OWF48060.1 GDHF01000564 JAI51750.1 GAKP01022214 JAC36738.1 KB632197 ERL90026.1 CH919640 EDW04562.1 APGK01024142 KB740539 ENN80440.1 PYGN01001816 PSN32681.1 GECZ01016435 GECZ01000289 JAS53334.1 JAS69480.1 NEVH01013550 PNF28809.1 KL216956 KFV72172.1 DQ215800 DQ215802 ACH45278.1 GEBQ01022706 GEBQ01014530 JAT17271.1 JAT25447.1 KV584048 OPL33240.1 MUZQ01000107 OWK58127.1 QUSF01000110 RLV91533.1 ABQF01034927 AEMK02000041 JH880804 ELR58733.1 BC105472 DQ215799 ACH45277.1 GL732552 EFX79310.1 KK843955 KFP85880.1
PZC74535.1 KQ459838 KPJ19717.1 GAIX01014109 JAA78451.1 EF619926 ABR09538.1 ODYU01012760 SOQ59229.1 AGBW02014044 OWR42353.1 KQ459460 KPJ00805.1 AXCM01007145 AXCN02001700 ATLV01012340 KE524783 KFB36510.1 DS231894 EDS44251.1 ADMH02001912 ETN60531.1 GFDL01015752 JAV19293.1 CH477650 EAT37789.1 AAAB01008816 EAA05187.5 JXUM01133026 KQ567923 KXJ69230.1 APCN01005461 CH963846 EDW72551.1 CH954177 EDV58201.1 CM002911 KMY92588.1 AF139064 AE013599 AY071443 CM000157 EDW89562.2 CH902619 EDV36805.1 CM000362 EDX06404.1 CH676472 EDW49402.1 CCAG010000346 CH933808 EDW09855.1 JXJN01011105 CVRI01000059 CRL03359.1 CH940648 EDW61115.1 CP012524 ALC42424.1 UFQT01000040 SSX18611.1 OUUW01000001 SPP74990.1 AJWK01020294 CH479210 EDW32924.1 CM000071 EDY69475.1 CH916367 EDW01241.1 QCYY01001403 ROT78273.1 NEDP02003738 OWF48060.1 GDHF01000564 JAI51750.1 GAKP01022214 JAC36738.1 KB632197 ERL90026.1 CH919640 EDW04562.1 APGK01024142 KB740539 ENN80440.1 PYGN01001816 PSN32681.1 GECZ01016435 GECZ01000289 JAS53334.1 JAS69480.1 NEVH01013550 PNF28809.1 KL216956 KFV72172.1 DQ215800 DQ215802 ACH45278.1 GEBQ01022706 GEBQ01014530 JAT17271.1 JAT25447.1 KV584048 OPL33240.1 MUZQ01000107 OWK58127.1 QUSF01000110 RLV91533.1 ABQF01034927 AEMK02000041 JH880804 ELR58733.1 BC105472 DQ215799 ACH45277.1 GL732552 EFX79310.1 KK843955 KFP85880.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000007151
UP000053268
UP000075881
+ More
UP000075883 UP000075886 UP000030765 UP000075920 UP000075880 UP000075885 UP000002320 UP000000673 UP000076408 UP000008820 UP000007062 UP000076407 UP000075903 UP000075884 UP000075900 UP000069940 UP000249989 UP000069272 UP000075840 UP000075902 UP000007798 UP000095301 UP000008711 UP000075882 UP000000803 UP000192221 UP000002282 UP000007801 UP000000304 UP000095300 UP000001292 UP000092444 UP000092445 UP000009192 UP000078200 UP000092443 UP000092460 UP000091820 UP000183832 UP000008792 UP000092553 UP000268350 UP000092461 UP000008744 UP000001819 UP000001070 UP000192223 UP000283509 UP000242188 UP000030742 UP000019118 UP000245037 UP000235965 UP000053875 UP000261681 UP000197619 UP000276834 UP000007754 UP000008227 UP000252040 UP000079721 UP000189705 UP000009136 UP000248480 UP000000305 UP000233160 UP000002280 UP000002279
UP000075883 UP000075886 UP000030765 UP000075920 UP000075880 UP000075885 UP000002320 UP000000673 UP000076408 UP000008820 UP000007062 UP000076407 UP000075903 UP000075884 UP000075900 UP000069940 UP000249989 UP000069272 UP000075840 UP000075902 UP000007798 UP000095301 UP000008711 UP000075882 UP000000803 UP000192221 UP000002282 UP000007801 UP000000304 UP000095300 UP000001292 UP000092444 UP000092445 UP000009192 UP000078200 UP000092443 UP000092460 UP000091820 UP000183832 UP000008792 UP000092553 UP000268350 UP000092461 UP000008744 UP000001819 UP000001070 UP000192223 UP000283509 UP000242188 UP000030742 UP000019118 UP000245037 UP000235965 UP000053875 UP000261681 UP000197619 UP000276834 UP000007754 UP000008227 UP000252040 UP000079721 UP000189705 UP000009136 UP000248480 UP000000305 UP000233160 UP000002280 UP000002279
Interpro
IPR008721
ORC6
+ More
IPR020529 ORC6_met/pln
IPR014710 RmlC-like_jellyroll
IPR018490 cNMP-bd-like
IPR018422 Cation/H_exchanger_CPA1
IPR005821 Ion_trans_dom
IPR027359 Volt_channel_dom_sf
IPR011993 PH-like_dom_sf
IPR039924 ICln/Lot5
IPR036812 NADP_OxRdtase_dom_sf
IPR020471 Aldo/keto_reductase
IPR023210 NADP_OxRdtase_dom
IPR018170 Aldo/ket_reductase_CS
IPR036915 Cyclin-like_sf
IPR020529 ORC6_met/pln
IPR014710 RmlC-like_jellyroll
IPR018490 cNMP-bd-like
IPR018422 Cation/H_exchanger_CPA1
IPR005821 Ion_trans_dom
IPR027359 Volt_channel_dom_sf
IPR011993 PH-like_dom_sf
IPR039924 ICln/Lot5
IPR036812 NADP_OxRdtase_dom_sf
IPR020471 Aldo/keto_reductase
IPR023210 NADP_OxRdtase_dom
IPR018170 Aldo/ket_reductase_CS
IPR036915 Cyclin-like_sf
Gene 3D
ProteinModelPortal
H9J431
D3VW61
A0A2A4K691
A0A2W1BLJ8
A0A0N1PIA9
S4NIE2
+ More
A6N831 A0A2H1X1P7 A0A212ELK0 A0A194Q728 A0A182K810 A0A182M6L8 A0A182Q437 A0A084VEW6 A0A182WDB5 A0A182IK57 A0A182P353 B0WD73 W5J968 A0A182YE87 A0A1Q3EVG5 Q16TG1 A0A1S4FQ03 Q7PSQ8 A0A182WR78 A0A182VFA2 A0A182MXF9 A0A182RXM7 A0A182HEY2 A0A182FUW9 A0A182HG20 A0A182TY94 B4MKB2 A0A1I8M4H2 B3N6V2 A0A182LHI8 A0A0J9R928 Q9Y1B2 A0A1W4UZ81 B4NWL7 B3MGP7 B4QHS0 A0A1I8P103 B4INL9 A0A1B0FMY7 A0A1A9ZD43 B4KMG7 A0A1A9UYX7 A0A1A9XLI1 A0A1B0BAQ1 A0A1A9W741 A0A1J1IT23 B4LNG3 A0A0M5IXY8 A0A336LP92 A0A3B0JHX9 A0A1B0GJG5 B4H588 B5E187 B4J8A4 A0A1W4XKD0 A0A3R7N652 A0A210QH20 A0A0K8WLN3 A0A034V5D3 U4UHE6 B4K2P2 N6UFC8 A0A2P8XL04 A0A1B6FT33 A0A2J7QJQ8 A0A093GZB4 B5G1W7 A0A1B6LPB1 A0A3L5TTG3 A0A383YXV5 A0A218UWG5 A0A3L8RZK0 H0ZEY5 I3LLV3 A0A341BYV8 A0A1S3ARQ1 A0A1U7SEN7 L8IRR5 Q2HJF3 B5G1W6 A0A2Y9DBW2 E9GM69 A0A2K6EW96 A0A091P2T6 F7EFU1 F7E7B7
A6N831 A0A2H1X1P7 A0A212ELK0 A0A194Q728 A0A182K810 A0A182M6L8 A0A182Q437 A0A084VEW6 A0A182WDB5 A0A182IK57 A0A182P353 B0WD73 W5J968 A0A182YE87 A0A1Q3EVG5 Q16TG1 A0A1S4FQ03 Q7PSQ8 A0A182WR78 A0A182VFA2 A0A182MXF9 A0A182RXM7 A0A182HEY2 A0A182FUW9 A0A182HG20 A0A182TY94 B4MKB2 A0A1I8M4H2 B3N6V2 A0A182LHI8 A0A0J9R928 Q9Y1B2 A0A1W4UZ81 B4NWL7 B3MGP7 B4QHS0 A0A1I8P103 B4INL9 A0A1B0FMY7 A0A1A9ZD43 B4KMG7 A0A1A9UYX7 A0A1A9XLI1 A0A1B0BAQ1 A0A1A9W741 A0A1J1IT23 B4LNG3 A0A0M5IXY8 A0A336LP92 A0A3B0JHX9 A0A1B0GJG5 B4H588 B5E187 B4J8A4 A0A1W4XKD0 A0A3R7N652 A0A210QH20 A0A0K8WLN3 A0A034V5D3 U4UHE6 B4K2P2 N6UFC8 A0A2P8XL04 A0A1B6FT33 A0A2J7QJQ8 A0A093GZB4 B5G1W7 A0A1B6LPB1 A0A3L5TTG3 A0A383YXV5 A0A218UWG5 A0A3L8RZK0 H0ZEY5 I3LLV3 A0A341BYV8 A0A1S3ARQ1 A0A1U7SEN7 L8IRR5 Q2HJF3 B5G1W6 A0A2Y9DBW2 E9GM69 A0A2K6EW96 A0A091P2T6 F7EFU1 F7E7B7
Ontologies
GO
Topology
Subcellular location
Nucleus
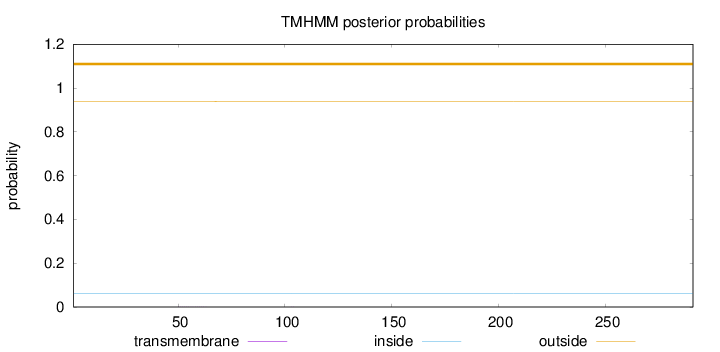
Length:
291
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00631
Exp number, first 60 AAs:
0.00257
Total prob of N-in:
0.06191
outside
1 - 291
Population Genetic Test Statistics
Pi
176.330584
Theta
140.950883
Tajima's D
0.583847
CLR
0.005729
CSRT
0.539423028848558
Interpretation
Uncertain
