Gene
KWMTBOMO12101 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA004343
Annotation
death-related_protein_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.137
Sequence
CDS
ATGTCGTCAACCGAGGAAACTTCACAACTAAAAGCTGGACATCCTCCTGCAGTAAAAGCTGGAGGTATGAGGATCACCCAACATAAAACTCCTCACTCAAAAGACAGCAAAGAACCAGCCAATGAAGACTTGACTGGTCTCTCTGGGCCTTCTCCCGTTCCGTCCAATCCGATCTCCATTGCTGGAGCTCCAAACAAAGGAACTAAGAGCAATATACAAAATAAAATGCTCTAA
Protein
MSSTEETSQLKAGHPPAVKAGGMRITQHKTPHSKDSKEPANEDLTGLSGPSPVPSNPISIAGAPNKGTKSNIQNKML
Summary
Uniprot
B8XWD7
A0A2W1BEC8
A0A2A4J829
A0A194QBJ3
I4DPI3
A0A0N1IIF7
+ More
A0A2H1VJD5 A0A1E1WRG2 S4PXP7 A0A0L7K377 A0A212EW74 A0A1I8Q8L3 A0A1I8N6C7 A0A1L8EHB6 A0A0K8TQ59 F4WW97 A0A0L0BNL8 A0A151XCR3 A0A195E7B0 A0A195B258 A0A158NLG4 A0A195F1S7 A0A0J7KX40 E2AA24 A0A154PQF6 E2C477 A0A026WZ74 D3TMX4 A0A0K8R5J7 A0A023F990 A0A2P8XWL1 A0A195CZU9 A0A1B0C6G1 A0A1A9WSG1 B4KTW5 R4WDV7 B4JVW2 A0A224XQF5 A0A069DVH0 B4LJ19 A0A0V0G383 V9IIA9 A0A087ZZ63 A0A023F871 A0A1W4UGE4 B4HNG6 B3NSH2 B4NSD9 Q7JRC0 B4P6I2 A0A2I2V0L7 A0A0P4VUM0 T1HQS3 A0A0M5J2C2 H3A5A4 A0A131Z871 L7M346 B3MFA6 A0A310ST26 A0A023FGF2 A0A286ZQE9 A0A023GDL1 G3MLP5 A0A2G8JYY7 A0A0L7RE43 A0A1S3DJB6 B4MP50 R4FPD7 A0A293MWD3 U4UKT6 Q28Y19 B4GGF7 A0A182UNU9 A0A182UA86 A0A182XGS7 A0NGR2 A0A3B0JL98 A0A087TBF0 Q6XIH1 A0A2Y9K3N8 G1LAM2 A2I444
A0A2H1VJD5 A0A1E1WRG2 S4PXP7 A0A0L7K377 A0A212EW74 A0A1I8Q8L3 A0A1I8N6C7 A0A1L8EHB6 A0A0K8TQ59 F4WW97 A0A0L0BNL8 A0A151XCR3 A0A195E7B0 A0A195B258 A0A158NLG4 A0A195F1S7 A0A0J7KX40 E2AA24 A0A154PQF6 E2C477 A0A026WZ74 D3TMX4 A0A0K8R5J7 A0A023F990 A0A2P8XWL1 A0A195CZU9 A0A1B0C6G1 A0A1A9WSG1 B4KTW5 R4WDV7 B4JVW2 A0A224XQF5 A0A069DVH0 B4LJ19 A0A0V0G383 V9IIA9 A0A087ZZ63 A0A023F871 A0A1W4UGE4 B4HNG6 B3NSH2 B4NSD9 Q7JRC0 B4P6I2 A0A2I2V0L7 A0A0P4VUM0 T1HQS3 A0A0M5J2C2 H3A5A4 A0A131Z871 L7M346 B3MFA6 A0A310ST26 A0A023FGF2 A0A286ZQE9 A0A023GDL1 G3MLP5 A0A2G8JYY7 A0A0L7RE43 A0A1S3DJB6 B4MP50 R4FPD7 A0A293MWD3 U4UKT6 Q28Y19 B4GGF7 A0A182UNU9 A0A182UA86 A0A182XGS7 A0NGR2 A0A3B0JL98 A0A087TBF0 Q6XIH1 A0A2Y9K3N8 G1LAM2 A2I444
Pubmed
19121390
28756777
26354079
22651552
23622113
26227816
+ More
22118469 25315136 26369729 21719571 26108605 21347285 20798317 24508170 30249741 20353571 25474469 29403074 17994087 23691247 26334808 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 17975172 27129103 9215903 26830274 25576852 22216098 29023486 23537049 15632085 12364791 14747013 17210077 14525923 20010809
22118469 25315136 26369729 21719571 26108605 21347285 20798317 24508170 30249741 20353571 25474469 29403074 17994087 23691247 26334808 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 17975172 27129103 9215903 26830274 25576852 22216098 29023486 23537049 15632085 12364791 14747013 17210077 14525923 20010809
EMBL
BABH01032647
FJ447481
ACJ38715.1
KZ150212
PZC72175.1
NWSH01002584
+ More
PCG67916.1 KQ459460 KPJ00806.1 AK403619 BAM19823.1 KQ459838 KPJ19713.1 ODYU01002891 SOQ40945.1 GDQN01001476 JAT89578.1 GAIX01004668 JAA87892.1 JTDY01012728 KOB52236.1 AGBW02012082 OWR45704.1 GFDG01000718 JAV18081.1 GDAI01001327 JAI16276.1 GL888404 EGI61568.1 JRES01001589 KNC21685.1 KQ982314 KYQ58068.1 KQ979568 KYN20724.1 KQ976662 KYM78370.1 ADTU01019398 KQ981864 KYN34418.1 LBMM01002422 KMQ94844.1 GL438001 EFN69723.1 KQ435037 KZC14129.1 GL452426 EFN77248.1 KK107063 QOIP01000005 EZA61046.1 RLU23125.1 CCAG010009508 EZ422776 ADD19052.1 GADI01007675 JAA66133.1 GBBI01001148 JAC17564.1 PYGN01001238 PSN36392.1 KQ977115 KYN05659.1 JXJN01026602 CH933808 EDW10691.1 AK417894 BAN21109.1 CH916375 EDV98100.1 GFTR01001690 JAW14736.1 GBGD01003580 JAC85309.1 CH940648 EDW60469.1 GECL01003548 JAP02576.1 JR046645 AEY60226.1 GBBI01001196 JAC17516.1 CH480816 EDW47402.1 CH954179 EDV56474.1 CH982175 CM002911 EDX15517.1 KMY92986.1 AE013599 BT001778 BT011048 KX532081 AAF58654.2 AAN71533.1 AAR31119.1 ANY27891.1 CM000158 EDW90934.1 AANG04003171 GDKW01000196 JAI56399.1 ACPB03022141 CP012524 ALC40774.1 AFYH01173130 AFYH01173131 AFYH01173132 AFYH01173133 AFYH01173134 AFYH01173135 GEDV01001901 JAP86656.1 GACK01006559 JAA58475.1 CH902619 EDV35580.1 KQ760243 OAD61275.1 GBBK01004403 JAC20079.1 AEMK02000102 GBBM01003157 JAC32261.1 JO842796 AEO34413.1 MRZV01001068 PIK40953.1 KQ414613 KOC69085.1 CH963848 EDW73889.1 GAHY01000934 JAA76576.1 GFWV01018747 MAA43475.1 KB632370 ERL93727.1 CM000071 EAL26147.1 CH479183 EDW35577.1 AAAB01008986 EAU75858.1 OUUW01000001 SPP74016.1 KK114440 KFM62439.1 AY231859 AAR09882.1 ACTA01106891 ACTA01114891 ACTA01122891 ACTA01130891 ACTA01138891 EF070547 ABM55613.1
PCG67916.1 KQ459460 KPJ00806.1 AK403619 BAM19823.1 KQ459838 KPJ19713.1 ODYU01002891 SOQ40945.1 GDQN01001476 JAT89578.1 GAIX01004668 JAA87892.1 JTDY01012728 KOB52236.1 AGBW02012082 OWR45704.1 GFDG01000718 JAV18081.1 GDAI01001327 JAI16276.1 GL888404 EGI61568.1 JRES01001589 KNC21685.1 KQ982314 KYQ58068.1 KQ979568 KYN20724.1 KQ976662 KYM78370.1 ADTU01019398 KQ981864 KYN34418.1 LBMM01002422 KMQ94844.1 GL438001 EFN69723.1 KQ435037 KZC14129.1 GL452426 EFN77248.1 KK107063 QOIP01000005 EZA61046.1 RLU23125.1 CCAG010009508 EZ422776 ADD19052.1 GADI01007675 JAA66133.1 GBBI01001148 JAC17564.1 PYGN01001238 PSN36392.1 KQ977115 KYN05659.1 JXJN01026602 CH933808 EDW10691.1 AK417894 BAN21109.1 CH916375 EDV98100.1 GFTR01001690 JAW14736.1 GBGD01003580 JAC85309.1 CH940648 EDW60469.1 GECL01003548 JAP02576.1 JR046645 AEY60226.1 GBBI01001196 JAC17516.1 CH480816 EDW47402.1 CH954179 EDV56474.1 CH982175 CM002911 EDX15517.1 KMY92986.1 AE013599 BT001778 BT011048 KX532081 AAF58654.2 AAN71533.1 AAR31119.1 ANY27891.1 CM000158 EDW90934.1 AANG04003171 GDKW01000196 JAI56399.1 ACPB03022141 CP012524 ALC40774.1 AFYH01173130 AFYH01173131 AFYH01173132 AFYH01173133 AFYH01173134 AFYH01173135 GEDV01001901 JAP86656.1 GACK01006559 JAA58475.1 CH902619 EDV35580.1 KQ760243 OAD61275.1 GBBK01004403 JAC20079.1 AEMK02000102 GBBM01003157 JAC32261.1 JO842796 AEO34413.1 MRZV01001068 PIK40953.1 KQ414613 KOC69085.1 CH963848 EDW73889.1 GAHY01000934 JAA76576.1 GFWV01018747 MAA43475.1 KB632370 ERL93727.1 CM000071 EAL26147.1 CH479183 EDW35577.1 AAAB01008986 EAU75858.1 OUUW01000001 SPP74016.1 KK114440 KFM62439.1 AY231859 AAR09882.1 ACTA01106891 ACTA01114891 ACTA01122891 ACTA01130891 ACTA01138891 EF070547 ABM55613.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000037510
UP000007151
+ More
UP000095300 UP000095301 UP000007755 UP000037069 UP000075809 UP000078492 UP000078540 UP000005205 UP000078541 UP000036403 UP000000311 UP000076502 UP000008237 UP000053097 UP000279307 UP000092444 UP000245037 UP000078542 UP000092460 UP000091820 UP000009192 UP000001070 UP000008792 UP000005203 UP000192221 UP000001292 UP000008711 UP000000304 UP000000803 UP000002282 UP000011712 UP000015103 UP000092553 UP000008672 UP000007801 UP000008227 UP000230750 UP000053825 UP000079169 UP000007798 UP000030742 UP000001819 UP000008744 UP000075903 UP000075902 UP000076407 UP000007062 UP000268350 UP000054359 UP000248482 UP000008912
UP000095300 UP000095301 UP000007755 UP000037069 UP000075809 UP000078492 UP000078540 UP000005205 UP000078541 UP000036403 UP000000311 UP000076502 UP000008237 UP000053097 UP000279307 UP000092444 UP000245037 UP000078542 UP000092460 UP000091820 UP000009192 UP000001070 UP000008792 UP000005203 UP000192221 UP000001292 UP000008711 UP000000304 UP000000803 UP000002282 UP000011712 UP000015103 UP000092553 UP000008672 UP000007801 UP000008227 UP000230750 UP000053825 UP000079169 UP000007798 UP000030742 UP000001819 UP000008744 UP000075903 UP000075902 UP000076407 UP000007062 UP000268350 UP000054359 UP000248482 UP000008912
Pfam
PF15228 DAP
Interpro
IPR024130
DAP1/DAPL1
ProteinModelPortal
B8XWD7
A0A2W1BEC8
A0A2A4J829
A0A194QBJ3
I4DPI3
A0A0N1IIF7
+ More
A0A2H1VJD5 A0A1E1WRG2 S4PXP7 A0A0L7K377 A0A212EW74 A0A1I8Q8L3 A0A1I8N6C7 A0A1L8EHB6 A0A0K8TQ59 F4WW97 A0A0L0BNL8 A0A151XCR3 A0A195E7B0 A0A195B258 A0A158NLG4 A0A195F1S7 A0A0J7KX40 E2AA24 A0A154PQF6 E2C477 A0A026WZ74 D3TMX4 A0A0K8R5J7 A0A023F990 A0A2P8XWL1 A0A195CZU9 A0A1B0C6G1 A0A1A9WSG1 B4KTW5 R4WDV7 B4JVW2 A0A224XQF5 A0A069DVH0 B4LJ19 A0A0V0G383 V9IIA9 A0A087ZZ63 A0A023F871 A0A1W4UGE4 B4HNG6 B3NSH2 B4NSD9 Q7JRC0 B4P6I2 A0A2I2V0L7 A0A0P4VUM0 T1HQS3 A0A0M5J2C2 H3A5A4 A0A131Z871 L7M346 B3MFA6 A0A310ST26 A0A023FGF2 A0A286ZQE9 A0A023GDL1 G3MLP5 A0A2G8JYY7 A0A0L7RE43 A0A1S3DJB6 B4MP50 R4FPD7 A0A293MWD3 U4UKT6 Q28Y19 B4GGF7 A0A182UNU9 A0A182UA86 A0A182XGS7 A0NGR2 A0A3B0JL98 A0A087TBF0 Q6XIH1 A0A2Y9K3N8 G1LAM2 A2I444
A0A2H1VJD5 A0A1E1WRG2 S4PXP7 A0A0L7K377 A0A212EW74 A0A1I8Q8L3 A0A1I8N6C7 A0A1L8EHB6 A0A0K8TQ59 F4WW97 A0A0L0BNL8 A0A151XCR3 A0A195E7B0 A0A195B258 A0A158NLG4 A0A195F1S7 A0A0J7KX40 E2AA24 A0A154PQF6 E2C477 A0A026WZ74 D3TMX4 A0A0K8R5J7 A0A023F990 A0A2P8XWL1 A0A195CZU9 A0A1B0C6G1 A0A1A9WSG1 B4KTW5 R4WDV7 B4JVW2 A0A224XQF5 A0A069DVH0 B4LJ19 A0A0V0G383 V9IIA9 A0A087ZZ63 A0A023F871 A0A1W4UGE4 B4HNG6 B3NSH2 B4NSD9 Q7JRC0 B4P6I2 A0A2I2V0L7 A0A0P4VUM0 T1HQS3 A0A0M5J2C2 H3A5A4 A0A131Z871 L7M346 B3MFA6 A0A310ST26 A0A023FGF2 A0A286ZQE9 A0A023GDL1 G3MLP5 A0A2G8JYY7 A0A0L7RE43 A0A1S3DJB6 B4MP50 R4FPD7 A0A293MWD3 U4UKT6 Q28Y19 B4GGF7 A0A182UNU9 A0A182UA86 A0A182XGS7 A0NGR2 A0A3B0JL98 A0A087TBF0 Q6XIH1 A0A2Y9K3N8 G1LAM2 A2I444
Ontologies
PANTHER
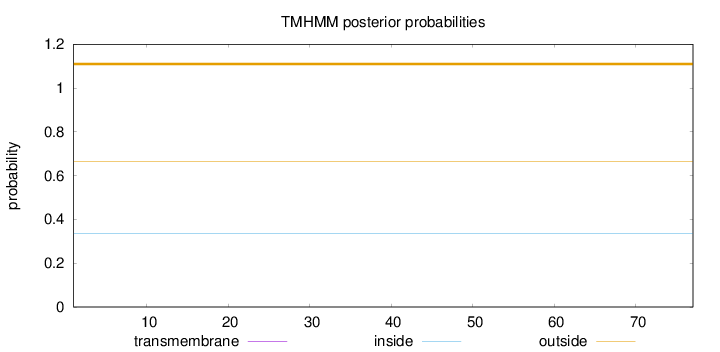
Topology
Length:
77
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0008
Exp number, first 60 AAs:
0.00065
Total prob of N-in:
0.33401
outside
1 - 77
Population Genetic Test Statistics
Pi
249.211078
Theta
197.296685
Tajima's D
0.728983
CLR
0.795247
CSRT
0.582470876456177
Interpretation
Uncertain
