Gene
KWMTBOMO12098 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA004340
Annotation
salivary_cysteine-rich_peptide_precursor_[Bombyx_mori]
Full name
NAD-dependent protein deacylase
Alternative Name
Regulatory protein SIR2 homolog 5
Location in the cell
Extracellular Reliability : 4.333
Sequence
CDS
ATGACATTCCTGTTGGGTGTGTTCGCGTTGGTTACCTTTGTCTTCTACGCCGAAGCCGCTGGCACGTGCCCTCTTCCTTCGAAAGTGTACGGTTGCAGCCCTAAATGCAAGGAGGACTACGAATGTACACACGGCAAGGTCTGCTGTTCTAATTCGTGCAACGCCAAGTCGTGTTCAGAGCCGGCTGCTTACGGCGGAGGCACCGGGAGCTCCAGCAAACACGCAACCGGCGGAACGGGCGTTTACTGCGAAAACGTCAAGTGTAACTCTTACGAAATATGCAAGCGTGATCCAATGACGAAGAGAATGAAATGCAGCAGAGCTTAA
Protein
MTFLLGVFALVTFVFYAEAAGTCPLPSKVYGCSPKCKEDYECTHGKVCCSNSCNAKSCSEPAAYGGGTGSSSKHATGGTGVYCENVKCNSYEICKRDPMTKRMKCSRA
Summary
Description
NAD-dependent lysine demalonylase, desuccinylase and deglutarylase that specifically removes malonyl, succinyl and glutaryl groups on target proteins. Has weak NAD-dependent protein deacetylase activity; however this activity may not be physiologically relevant in vivo.
Catalytic Activity
H2O + N(6)-glutaryl-L-lysyl-[protein] + NAD(+) = 2''-O-glutaryl-ADP-D-ribose + L-lysyl-[protein] + nicotinamide
H2O + N(6)-malonyl-L-lysyl-[protein] + NAD(+) = 2''-O-malonyl-ADP-D-ribose + L-lysyl-[protein] + nicotinamide
H2O + N(6)-succinyl-L-lysyl-[protein] + NAD(+) = 2''-O-succinyl-ADP-D-ribose + L-lysyl-[protein] + nicotinamide
H2O + N(6)-malonyl-L-lysyl-[protein] + NAD(+) = 2''-O-malonyl-ADP-D-ribose + L-lysyl-[protein] + nicotinamide
H2O + N(6)-succinyl-L-lysyl-[protein] + NAD(+) = 2''-O-succinyl-ADP-D-ribose + L-lysyl-[protein] + nicotinamide
Cofactor
Zn(2+)
Similarity
Belongs to the sirtuin family. Class III subfamily.
Belongs to the peptidase S1 family.
Belongs to the peptidase S1 family.
Uniprot
H9J4A1
Q2F5L7
A0A2A4J4S4
A0A2H1WEN5
A0A1E1W0N8
I4DJI2
+ More
A0A194Q5I6 A0A0N0PEC3 I4DME0 A0A2W1AYL1 B0WGW5 Q6TS02 B8RJ27 A0A1I8NYZ9 B8RJ73 U5ESJ3 A0A2A4J4P9 A0A336L5S8 T1DJ53 A0A0T6AZZ9 B4MRJ9 B4J690 A0A1A9WC59 A0A1L8E9B7 A0A1L8E8X8 A0A1L8E9C8 A0A1B0BFS7 A0A1L8DP95 A0A1A9Y1T5 Q291D9 B4GAV0 A0A1L8E959 A0A1L8E9E2 A0A1L8E8W9 A0A1A9VSF7 A0A0K8VYK3 A0A1S3DI78 B4LLP0 A0A0L0CPL9 A0A0K8UBV8 A0A0M4EV16 T1H3T2 A0A0K8V321 A0A023EEW1 A0A023EF48 A0A1I8MRC7 A0A1L8E8L7 A0A034WMT9 A0A1I8M4C9 A0A023EF50 A0A0L0CPZ0 A0A0A1XAB4 A0A026WL57 N6UHB8 B3NR33 B4QFV2 A0A182V7J1 A0A182XC99 Q7Q332 A0A182HNK0 A0A0C9R9D0 A0A1W6EVZ4 B4P770 Q8MKJ5 B4KP03 A0A348G5X8 A0A1W4UHL7 A0A310SA66 D6X3Z4 A0A154PFW7 B4HRE2 A0A2A3E681 E2AH81 W5JSZ0 A0A182P9C4 A0A087ZR77 A0A2M3ZB73 W8C475 Q16PH2 A0A2P8ZKJ5 A0A1A9Z4T8 A0A2M4CZF3 B3ME19 A0A182QP64 A0A158NA96 W5JPJ3 A0A2M3ZBD0 A0A182J089 A0A182UHK7 A0A182YPF3 A0A182FTD9 A0A1Y1KSH3 A0A2M4ACT1 A0A2M4ACS1 A0A182WF61 A0A1I8JU61 A0A2M3YW39
A0A194Q5I6 A0A0N0PEC3 I4DME0 A0A2W1AYL1 B0WGW5 Q6TS02 B8RJ27 A0A1I8NYZ9 B8RJ73 U5ESJ3 A0A2A4J4P9 A0A336L5S8 T1DJ53 A0A0T6AZZ9 B4MRJ9 B4J690 A0A1A9WC59 A0A1L8E9B7 A0A1L8E8X8 A0A1L8E9C8 A0A1B0BFS7 A0A1L8DP95 A0A1A9Y1T5 Q291D9 B4GAV0 A0A1L8E959 A0A1L8E9E2 A0A1L8E8W9 A0A1A9VSF7 A0A0K8VYK3 A0A1S3DI78 B4LLP0 A0A0L0CPL9 A0A0K8UBV8 A0A0M4EV16 T1H3T2 A0A0K8V321 A0A023EEW1 A0A023EF48 A0A1I8MRC7 A0A1L8E8L7 A0A034WMT9 A0A1I8M4C9 A0A023EF50 A0A0L0CPZ0 A0A0A1XAB4 A0A026WL57 N6UHB8 B3NR33 B4QFV2 A0A182V7J1 A0A182XC99 Q7Q332 A0A182HNK0 A0A0C9R9D0 A0A1W6EVZ4 B4P770 Q8MKJ5 B4KP03 A0A348G5X8 A0A1W4UHL7 A0A310SA66 D6X3Z4 A0A154PFW7 B4HRE2 A0A2A3E681 E2AH81 W5JSZ0 A0A182P9C4 A0A087ZR77 A0A2M3ZB73 W8C475 Q16PH2 A0A2P8ZKJ5 A0A1A9Z4T8 A0A2M4CZF3 B3ME19 A0A182QP64 A0A158NA96 W5JPJ3 A0A2M3ZBD0 A0A182J089 A0A182UHK7 A0A182YPF3 A0A182FTD9 A0A1Y1KSH3 A0A2M4ACT1 A0A2M4ACS1 A0A182WF61 A0A1I8JU61 A0A2M3YW39
EC Number
3.5.1.-
Pubmed
19121390
22651552
26354079
28756777
24330624
17994087
+ More
15632085 26108605 24945155 25315136 25348373 26483478 25830018 24508170 30249741 23537049 22936249 12364791 14747013 17210077 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 18362917 19820115 20798317 20920257 23761445 24495485 17510324 29403074 21347285 25244985 28004739
15632085 26108605 24945155 25315136 25348373 26483478 25830018 24508170 30249741 23537049 22936249 12364791 14747013 17210077 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 18362917 19820115 20798317 20920257 23761445 24495485 17510324 29403074 21347285 25244985 28004739
EMBL
BABH01032641
DQ311406
ABD36350.1
NWSH01003389
PCG66392.1
ODYU01008163
+ More
SOQ51533.1 GDQN01010518 JAT80536.1 AK401450 BAM18072.1 KQ459460 KPJ00808.1 KQ459838 KPJ19711.1 AK402458 BAM19080.1 KZ150735 PZC70352.1 DS231930 EDS27259.1 AY388547 DS233801 AAR18435.1 EDS31649.1 EZ000452 ACJ64326.1 EZ000498 ACJ64372.1 GANO01004810 JAB55061.1 PCG66393.1 UFQS01001098 UFQT01001098 SSX09000.1 SSX28911.1 GALA01000506 JAA94346.1 LJIG01016430 KRT80626.1 CH963850 EDW74738.2 CH916367 EDW00863.1 GFDG01003590 JAV15209.1 GFDG01003592 JAV15207.1 GFDG01003593 JAV15206.1 JXJN01013644 JXJN01013645 JXJN01013646 GFDF01005887 JAV08197.1 CM000071 EAL25173.2 CH479181 EDW32052.1 GFDG01003595 JAV15204.1 GFDG01003594 JAV15205.1 GFDG01003591 JAV15208.1 GDHF01019816 GDHF01008372 JAI32498.1 JAI43942.1 CH940648 EDW60903.2 JRES01000080 KNC34323.1 GDHF01028162 JAI24152.1 CP012524 ALC41637.1 CAQQ02071450 GDHF01019088 JAI33226.1 GAPW01006208 GAPW01006207 GAPW01006206 GAPW01006205 JAC07391.1 GAPW01006209 JAC07389.1 GFDG01003702 JAV15097.1 GAKP01002983 JAC55969.1 JXUM01122149 GAPW01006204 KQ566574 JAC07394.1 KXJ70015.1 KNC34331.1 GBXI01006679 JAD07613.1 KK107167 QOIP01000010 EZA56401.1 RLU18038.1 APGK01035417 KB740925 KB631924 ENN78032.1 ERL87273.1 CH954179 EDV56022.1 CM000362 CM002911 EDX07097.1 KMY93783.1 AAAB01008966 EAA12985.4 APCN01001938 GBYB01013009 JAG82776.1 KY563487 ARK19896.1 CM000158 EDW91035.1 AY113384 AE013599 KX531668 AAM29389.1 AAM68554.1 ANY27478.1 CH933808 EDW10069.1 FX985515 BBF97851.1 KQ765605 OAD53818.1 KQ971379 EEZ97346.1 KQ434896 KZC10759.1 CH480816 EDW47871.1 KZ288375 PBC26679.1 GL439483 EFN67198.1 ADMH02000540 ETN66045.1 GGFM01005000 MBW25751.1 GAMC01002442 JAC04114.1 CH477781 EAT36276.1 PYGN01000028 PSN57021.1 GGFL01006509 MBW70687.1 CH902619 EDV37564.2 AXCN02002114 ADTU01010081 ETN66046.1 GGFM01005034 MBW25785.1 GEZM01075029 JAV64333.1 GGFK01005258 MBW38579.1 GGFK01005248 MBW38569.1 GGFF01000001 MBW20468.1
SOQ51533.1 GDQN01010518 JAT80536.1 AK401450 BAM18072.1 KQ459460 KPJ00808.1 KQ459838 KPJ19711.1 AK402458 BAM19080.1 KZ150735 PZC70352.1 DS231930 EDS27259.1 AY388547 DS233801 AAR18435.1 EDS31649.1 EZ000452 ACJ64326.1 EZ000498 ACJ64372.1 GANO01004810 JAB55061.1 PCG66393.1 UFQS01001098 UFQT01001098 SSX09000.1 SSX28911.1 GALA01000506 JAA94346.1 LJIG01016430 KRT80626.1 CH963850 EDW74738.2 CH916367 EDW00863.1 GFDG01003590 JAV15209.1 GFDG01003592 JAV15207.1 GFDG01003593 JAV15206.1 JXJN01013644 JXJN01013645 JXJN01013646 GFDF01005887 JAV08197.1 CM000071 EAL25173.2 CH479181 EDW32052.1 GFDG01003595 JAV15204.1 GFDG01003594 JAV15205.1 GFDG01003591 JAV15208.1 GDHF01019816 GDHF01008372 JAI32498.1 JAI43942.1 CH940648 EDW60903.2 JRES01000080 KNC34323.1 GDHF01028162 JAI24152.1 CP012524 ALC41637.1 CAQQ02071450 GDHF01019088 JAI33226.1 GAPW01006208 GAPW01006207 GAPW01006206 GAPW01006205 JAC07391.1 GAPW01006209 JAC07389.1 GFDG01003702 JAV15097.1 GAKP01002983 JAC55969.1 JXUM01122149 GAPW01006204 KQ566574 JAC07394.1 KXJ70015.1 KNC34331.1 GBXI01006679 JAD07613.1 KK107167 QOIP01000010 EZA56401.1 RLU18038.1 APGK01035417 KB740925 KB631924 ENN78032.1 ERL87273.1 CH954179 EDV56022.1 CM000362 CM002911 EDX07097.1 KMY93783.1 AAAB01008966 EAA12985.4 APCN01001938 GBYB01013009 JAG82776.1 KY563487 ARK19896.1 CM000158 EDW91035.1 AY113384 AE013599 KX531668 AAM29389.1 AAM68554.1 ANY27478.1 CH933808 EDW10069.1 FX985515 BBF97851.1 KQ765605 OAD53818.1 KQ971379 EEZ97346.1 KQ434896 KZC10759.1 CH480816 EDW47871.1 KZ288375 PBC26679.1 GL439483 EFN67198.1 ADMH02000540 ETN66045.1 GGFM01005000 MBW25751.1 GAMC01002442 JAC04114.1 CH477781 EAT36276.1 PYGN01000028 PSN57021.1 GGFL01006509 MBW70687.1 CH902619 EDV37564.2 AXCN02002114 ADTU01010081 ETN66046.1 GGFM01005034 MBW25785.1 GEZM01075029 JAV64333.1 GGFK01005258 MBW38579.1 GGFK01005248 MBW38569.1 GGFF01000001 MBW20468.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000002320
UP000095300
+ More
UP000007798 UP000001070 UP000091820 UP000092460 UP000092443 UP000001819 UP000008744 UP000078200 UP000079169 UP000008792 UP000037069 UP000092553 UP000015102 UP000095301 UP000069940 UP000249989 UP000053097 UP000279307 UP000019118 UP000030742 UP000008711 UP000000304 UP000075903 UP000076407 UP000007062 UP000075840 UP000002282 UP000000803 UP000009192 UP000192221 UP000007266 UP000076502 UP000001292 UP000242457 UP000000311 UP000000673 UP000075885 UP000005203 UP000008820 UP000245037 UP000092445 UP000007801 UP000075886 UP000005205 UP000075880 UP000075902 UP000076408 UP000069272 UP000075920 UP000075900
UP000007798 UP000001070 UP000091820 UP000092460 UP000092443 UP000001819 UP000008744 UP000078200 UP000079169 UP000008792 UP000037069 UP000092553 UP000015102 UP000095301 UP000069940 UP000249989 UP000053097 UP000279307 UP000019118 UP000030742 UP000008711 UP000000304 UP000075903 UP000076407 UP000007062 UP000075840 UP000002282 UP000000803 UP000009192 UP000192221 UP000007266 UP000076502 UP000001292 UP000242457 UP000000311 UP000000673 UP000075885 UP000005203 UP000008820 UP000245037 UP000092445 UP000007801 UP000075886 UP000005205 UP000075880 UP000075902 UP000076408 UP000069272 UP000075920 UP000075900
Interpro
Gene 3D
ProteinModelPortal
H9J4A1
Q2F5L7
A0A2A4J4S4
A0A2H1WEN5
A0A1E1W0N8
I4DJI2
+ More
A0A194Q5I6 A0A0N0PEC3 I4DME0 A0A2W1AYL1 B0WGW5 Q6TS02 B8RJ27 A0A1I8NYZ9 B8RJ73 U5ESJ3 A0A2A4J4P9 A0A336L5S8 T1DJ53 A0A0T6AZZ9 B4MRJ9 B4J690 A0A1A9WC59 A0A1L8E9B7 A0A1L8E8X8 A0A1L8E9C8 A0A1B0BFS7 A0A1L8DP95 A0A1A9Y1T5 Q291D9 B4GAV0 A0A1L8E959 A0A1L8E9E2 A0A1L8E8W9 A0A1A9VSF7 A0A0K8VYK3 A0A1S3DI78 B4LLP0 A0A0L0CPL9 A0A0K8UBV8 A0A0M4EV16 T1H3T2 A0A0K8V321 A0A023EEW1 A0A023EF48 A0A1I8MRC7 A0A1L8E8L7 A0A034WMT9 A0A1I8M4C9 A0A023EF50 A0A0L0CPZ0 A0A0A1XAB4 A0A026WL57 N6UHB8 B3NR33 B4QFV2 A0A182V7J1 A0A182XC99 Q7Q332 A0A182HNK0 A0A0C9R9D0 A0A1W6EVZ4 B4P770 Q8MKJ5 B4KP03 A0A348G5X8 A0A1W4UHL7 A0A310SA66 D6X3Z4 A0A154PFW7 B4HRE2 A0A2A3E681 E2AH81 W5JSZ0 A0A182P9C4 A0A087ZR77 A0A2M3ZB73 W8C475 Q16PH2 A0A2P8ZKJ5 A0A1A9Z4T8 A0A2M4CZF3 B3ME19 A0A182QP64 A0A158NA96 W5JPJ3 A0A2M3ZBD0 A0A182J089 A0A182UHK7 A0A182YPF3 A0A182FTD9 A0A1Y1KSH3 A0A2M4ACT1 A0A2M4ACS1 A0A182WF61 A0A1I8JU61 A0A2M3YW39
A0A194Q5I6 A0A0N0PEC3 I4DME0 A0A2W1AYL1 B0WGW5 Q6TS02 B8RJ27 A0A1I8NYZ9 B8RJ73 U5ESJ3 A0A2A4J4P9 A0A336L5S8 T1DJ53 A0A0T6AZZ9 B4MRJ9 B4J690 A0A1A9WC59 A0A1L8E9B7 A0A1L8E8X8 A0A1L8E9C8 A0A1B0BFS7 A0A1L8DP95 A0A1A9Y1T5 Q291D9 B4GAV0 A0A1L8E959 A0A1L8E9E2 A0A1L8E8W9 A0A1A9VSF7 A0A0K8VYK3 A0A1S3DI78 B4LLP0 A0A0L0CPL9 A0A0K8UBV8 A0A0M4EV16 T1H3T2 A0A0K8V321 A0A023EEW1 A0A023EF48 A0A1I8MRC7 A0A1L8E8L7 A0A034WMT9 A0A1I8M4C9 A0A023EF50 A0A0L0CPZ0 A0A0A1XAB4 A0A026WL57 N6UHB8 B3NR33 B4QFV2 A0A182V7J1 A0A182XC99 Q7Q332 A0A182HNK0 A0A0C9R9D0 A0A1W6EVZ4 B4P770 Q8MKJ5 B4KP03 A0A348G5X8 A0A1W4UHL7 A0A310SA66 D6X3Z4 A0A154PFW7 B4HRE2 A0A2A3E681 E2AH81 W5JSZ0 A0A182P9C4 A0A087ZR77 A0A2M3ZB73 W8C475 Q16PH2 A0A2P8ZKJ5 A0A1A9Z4T8 A0A2M4CZF3 B3ME19 A0A182QP64 A0A158NA96 W5JPJ3 A0A2M3ZBD0 A0A182J089 A0A182UHK7 A0A182YPF3 A0A182FTD9 A0A1Y1KSH3 A0A2M4ACT1 A0A2M4ACS1 A0A182WF61 A0A1I8JU61 A0A2M3YW39
Ontologies
GO
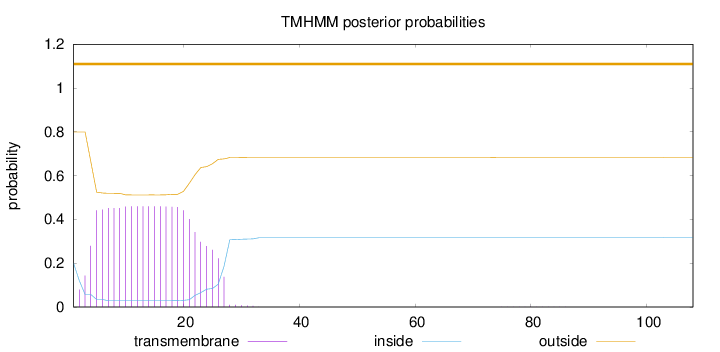
Topology
Subcellular location
Mitochondrion
SignalP
Position: 1 - 19,
Likelihood: 0.999794
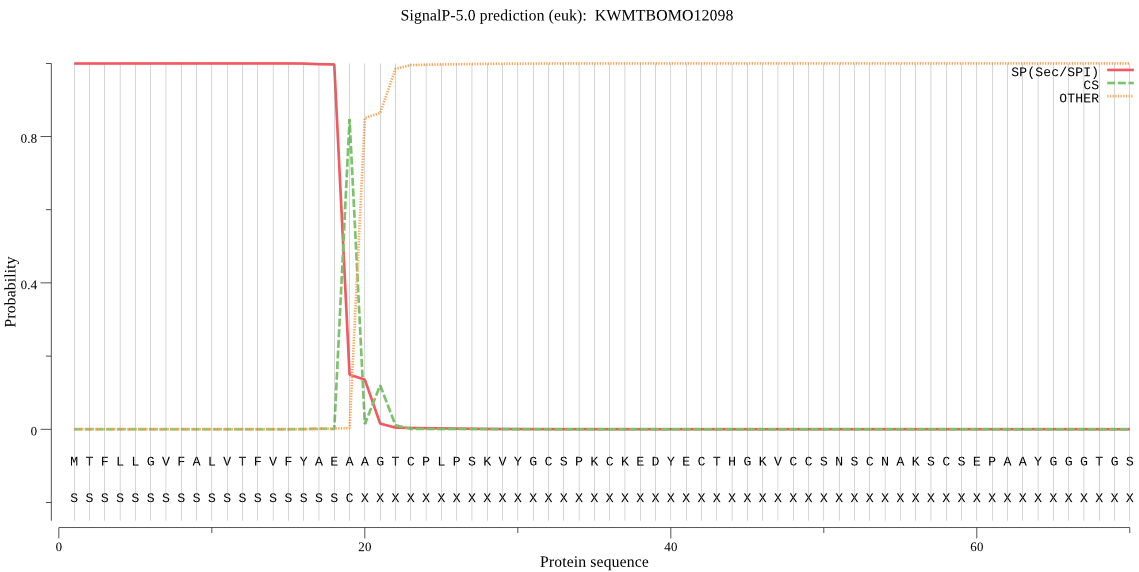
Length:
108
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
9.75621
Exp number, first 60 AAs:
9.75332
Total prob of N-in:
0.20062
outside
1 - 108
Population Genetic Test Statistics
Pi
22.258078
Theta
62.39927
Tajima's D
-1.951073
CLR
1.216461
CSRT
0.0184990750462477
Interpretation
Uncertain
