Gene
KWMTBOMO12090
Pre Gene Modal
BGIBMGA004338
Annotation
Uncharacterized_protein_OBRU01_14583_[Operophtera_brumata]
Location in the cell
Cytoplasmic Reliability : 1.451
Sequence
CDS
ATGGCTAGTAGCATTCATAATTACGGCGTCGAAGTGTCGGCTGAAGAAAAGGAATTTCAAAATGTTTGGAATAGATTTTCGGAATGGTTGCATTGTATATGTGTAGTAACATTCGACTTAGAGCTTGGTCAAGCAATGGAAAGTGTCTTCCCACCCGGAGTACCGTTGACTGATCAAGAAAAATGCAATATTTGTTATTTGGCGTTTCCGGATTCCAATTCAGGATGTATGGGGGACACCCAGTTTCATGTGCGGCTTCGCTCATTCATGTCCTTGACACCACAACAACTTGCATACAACGAAGATAGCGTGCCCACACTTCGCGCTGATTTAACACATTACTGGGGCTTCGTCTACTTTAGACAGGTCAAGGACTCTTCGTTACCAAGAGGTTACTTCCAAAAGAGCATAATCCTGCTGAGCCGACTGCCTTTCATCAATTTGTTCTACAAAGTTATTCAACTCATAGCTCCAAAACACTTCGAAGATGGTGAAAGTAGCTTGGAAGCTGCCTGTCATGATATAAATAGATGGCCTTCATTGAAGGCGGGCCAAAATGTACTGCTGCCAGTGTTGGGCACTGTGTTTCAGTCATACATTCCTAATCAACAAACTGGAAAAATTGTAAGGTCAGACATTGCAGAACAAATTCAATCACACAATGTACCTCATGTTGTTGCCTCTTTACAGGATGTTAATGTTTTTGATGCCTTATCTAGCTTAATCTCACATTTACATTTACTGTGGGAATTGGTTTTGACTGCTGAGCCAATTGTAGTGATGGCTAGCTCGCCAACCGAATGCTCTGCGTTGGTCCAAGCATTGACAAATTTAATCCAACCTCTCCCATATTCAGCAGAGTATAGACCATATTTCACAATACATGACAGTGAATTCAGAGAGTTCACAAGGAAACAATTCAACCCCCCCTGTGTAATACTGGGTGTGACTAATCCATTCTTCACAAAAACACTGCAACACTGGCCCCACACCATAAAACTTGGTGACGTCACTTCTATAAAAACGAAGCTGAAAAAAATTGGTAACTTGAAACTACTTGACACTGCTCCAGGTGTATACACTCAGTACAAAGCATTCTTAGAAAAAGATAAAGCAATTATAAAAAAACTTCACAACGGAATCCGTACTGAAAGGCCTTCGGAAGTTCAGACTGCAATGGTCAAAAGACATCTACTAGAACTGACTCAAAGCTTTATGATTCCTCTAGAGAGGTATATGGCATCACTCATGCCATTACAGAAAAACATTTCACCATTTAGAGCACCTCCTATACCCAATCCGTTCAACCCGGAAGACTTCTTTGCTATGTTACAGCAATCAGGACCTCAATTAACGACTGGCATTAAAGGTGACTGGATAGGATTGTACAGAAACTTTTTCAAAACCCCAAACTTCAGTGCTTGGTTTCACGAGAGACACAATAATTTGACAAACAAGCTACAAGCACTGCAGCTTGAAGCTTTAGCTGGTAGTGACCTGAAGCAATGGTCTAAAGGGAAGAAAGAAATTGAAATTGTTGATATGGTCCTCAAACTCAGGGAGGTGTTGAAAAGTAGTCCCCCAGTGCCTGACAATACAAAGACGTTGTTGGCAAGACGTCTTGATGATCTGAACTGTGTTTTGCCTGATGACATGAAATCTATATTGAATGCAGCCTCGTGA
Protein
MASSIHNYGVEVSAEEKEFQNVWNRFSEWLHCICVVTFDLELGQAMESVFPPGVPLTDQEKCNICYLAFPDSNSGCMGDTQFHVRLRSFMSLTPQQLAYNEDSVPTLRADLTHYWGFVYFRQVKDSSLPRGYFQKSIILLSRLPFINLFYKVIQLIAPKHFEDGESSLEAACHDINRWPSLKAGQNVLLPVLGTVFQSYIPNQQTGKIVRSDIAEQIQSHNVPHVVASLQDVNVFDALSSLISHLHLLWELVLTAEPIVVMASSPTECSALVQALTNLIQPLPYSAEYRPYFTIHDSEFREFTRKQFNPPCVILGVTNPFFTKTLQHWPHTIKLGDVTSIKTKLKKIGNLKLLDTAPGVYTQYKAFLEKDKAIIKKLHNGIRTERPSEVQTAMVKRHLLELTQSFMIPLERYMASLMPLQKNISPFRAPPIPNPFNPEDFFAMLQQSGPQLTTGIKGDWIGLYRNFFKTPNFSAWFHERHNNLTNKLQALQLEALAGSDLKQWSKGKKEIEIVDMVLKLREVLKSSPPVPDNTKTLLARRLDDLNCVLPDDMKSILNAAS
Summary
Similarity
Belongs to the heat shock protein 70 family.
Uniprot
H9J499
A0A2H1WWP8
A0A2A4JV49
A0A2W1BKG2
A0A0L7L6F2
A0A0N1IAN4
+ More
A0A194Q738 A0A212ESQ9 A0A067RD28 A0A0M8ZYX6 E2AYA4 A0A1B6E4R2 A0A3L8DS24 A0A195C8B5 E9IFT0 E0VCM1 A0A195BFA3 A0A195FE38 A0A158NBL2 A0A026W7X6 A0A195DR80 E2BIF2 F4X8J6 A0A1Q3FA67 A0A1Q3F9W8 A0A151WF39 A0A1Q3F997 A0A0J7KZC7 A0A1Y1N4D4 D6W6S7 T1IWF4 A0A0L7QRQ8 B0XHS2 A0A182GEX8 A0A1Y1N4B0 A0A0P6ISR5 K7J104 A0A232F6R2 A0A182RB36 A0A2S2R3J4 J9K9J3 A0A2R7WHG3 A0A3S1HPY0 A0A182P9Y8 A0A0T6AWD1 A0A1W4X400 A0A2A3E8P0 A0A088ALK5 A0A210QAF4 A0A0A9XNT0 A0A336MJK2 A0A336LZW9 A0A1S3J2X2 A0A1B6CG68 V3ZTW0 K1QS76 A0A1S3J3D7 A0A023F3J2 A0A224XAF2 T1HWP1 W4YH44 N6U6Q9 R7U310 A0A2J7PNA7 A0A0P4VZK7 A0A1D2MH41 A7RNP1 V5HT62 A0A2B4RXL3 A0A3P8VA71 W5NJE1 A0A1A8NQL7 A0A3B3SUS2 A0A1A8L7N8 A0A3Q3GN48 A0A3Q3QCX2 A0A3Q3QCV8 A0A1A8QAM4 A0A1E1XBT9 A0A1W5B026 A0A1B6KPM2 A0A1A8ARM9 A0A3B3VBL9 A0A3Q3GG73 A0A1A8RXV6 A0A1A8N3P1 A0A131XJ80 A0A3Q2E0H9 A0A146P3I3 A0A3P8Y692 A0A1A8DYT5 A0A3Q3A706 A0A3B4DNF4 A0A3B5B8J6 A0A2I4CVY7 A0A3B3DWI1 A0A1S3MYN6 A0A2C9LJC6 A0A1A8BDC5
A0A194Q738 A0A212ESQ9 A0A067RD28 A0A0M8ZYX6 E2AYA4 A0A1B6E4R2 A0A3L8DS24 A0A195C8B5 E9IFT0 E0VCM1 A0A195BFA3 A0A195FE38 A0A158NBL2 A0A026W7X6 A0A195DR80 E2BIF2 F4X8J6 A0A1Q3FA67 A0A1Q3F9W8 A0A151WF39 A0A1Q3F997 A0A0J7KZC7 A0A1Y1N4D4 D6W6S7 T1IWF4 A0A0L7QRQ8 B0XHS2 A0A182GEX8 A0A1Y1N4B0 A0A0P6ISR5 K7J104 A0A232F6R2 A0A182RB36 A0A2S2R3J4 J9K9J3 A0A2R7WHG3 A0A3S1HPY0 A0A182P9Y8 A0A0T6AWD1 A0A1W4X400 A0A2A3E8P0 A0A088ALK5 A0A210QAF4 A0A0A9XNT0 A0A336MJK2 A0A336LZW9 A0A1S3J2X2 A0A1B6CG68 V3ZTW0 K1QS76 A0A1S3J3D7 A0A023F3J2 A0A224XAF2 T1HWP1 W4YH44 N6U6Q9 R7U310 A0A2J7PNA7 A0A0P4VZK7 A0A1D2MH41 A7RNP1 V5HT62 A0A2B4RXL3 A0A3P8VA71 W5NJE1 A0A1A8NQL7 A0A3B3SUS2 A0A1A8L7N8 A0A3Q3GN48 A0A3Q3QCX2 A0A3Q3QCV8 A0A1A8QAM4 A0A1E1XBT9 A0A1W5B026 A0A1B6KPM2 A0A1A8ARM9 A0A3B3VBL9 A0A3Q3GG73 A0A1A8RXV6 A0A1A8N3P1 A0A131XJ80 A0A3Q2E0H9 A0A146P3I3 A0A3P8Y692 A0A1A8DYT5 A0A3Q3A706 A0A3B4DNF4 A0A3B5B8J6 A0A2I4CVY7 A0A3B3DWI1 A0A1S3MYN6 A0A2C9LJC6 A0A1A8BDC5
Pubmed
19121390
28756777
26227816
26354079
22118469
24845553
+ More
20798317 30249741 21282665 20566863 21347285 24508170 21719571 28004739 18362917 19820115 26483478 26999592 20075255 28648823 28812685 25401762 23254933 22992520 25474469 23537049 27289101 17615350 25765539 24487278 29240929 28503490 28049606 25069045 29451363 15562597
20798317 30249741 21282665 20566863 21347285 24508170 21719571 28004739 18362917 19820115 26483478 26999592 20075255 28648823 28812685 25401762 23254933 22992520 25474469 23537049 27289101 17615350 25765539 24487278 29240929 28503490 28049606 25069045 29451363 15562597
EMBL
BABH01032622
ODYU01011631
SOQ57495.1
NWSH01000561
PCG75646.1
KZ150212
+ More
PZC72163.1 JTDY01002701 KOB70889.1 KQ459838 KPJ19705.1 KQ459460 KPJ00815.1 AGBW02012739 OWR44520.1 KK852539 KDR21781.1 KQ435794 KOX73747.1 GL443836 EFN61587.1 GEDC01004387 JAS32911.1 QOIP01000004 RLU23230.1 KQ978219 KYM96411.1 GL762897 EFZ20569.1 DS235059 EEB11127.1 KQ976500 KYM83261.1 KQ981673 KYN38289.1 ADTU01011209 KK107372 EZA51746.1 KQ980581 KYN15351.1 GL448504 EFN84540.1 GL888932 EGI57265.1 GFDL01010618 JAV24427.1 GFDL01010732 JAV24313.1 KQ983227 KYQ46470.1 GFDL01010933 JAV24112.1 LBMM01001848 KMQ95653.1 GEZM01013114 JAV92742.1 KQ971307 EFA11536.1 JH431613 KQ414775 KOC61307.1 DS233195 EDS28591.1 JXUM01058934 KQ562026 KXJ76872.1 GEZM01013115 JAV92741.1 GDUN01001052 JAN94867.1 NNAY01000821 OXU26335.1 GGMS01014709 MBY83912.1 ABLF02033596 KK854831 PTY19087.1 RQTK01000228 RUS83833.1 LJIG01022709 KRT79118.1 KZ288324 PBC28095.1 NEDP02004420 OWF45695.1 GBHO01022308 GBRD01011615 GBRD01008524 JAG21296.1 JAG54209.1 UFQS01001027 UFQT01001027 SSX08621.1 SSX28537.1 UFQS01000371 UFQT01000371 SSX03260.1 SSX23626.1 GEDC01024887 JAS12411.1 KB203534 ESO84336.1 JH816704 EKC31765.1 GBBI01002821 JAC15891.1 GFTR01007121 JAW09305.1 ACPB03016234 AAGJ04114875 APGK01047232 KB741077 KB631792 ENN74252.1 ERL86121.1 AMQN01001815 KB305766 ELU00735.1 NEVH01023955 PNF17823.1 GDRN01100812 GDRN01100811 JAI58495.1 LJIJ01001303 ODM92184.1 DS469523 EDO46962.1 GANP01002939 JAB81529.1 LSMT01000283 PFX21300.1 AHAT01023625 HAEI01000345 HAEH01007472 SBR71330.1 HAEF01003116 SBR40498.1 HAEG01011855 SBR90830.1 GFAC01002607 JAT96581.1 GEBQ01026606 JAT13371.1 HADY01019312 SBP57797.1 HAEH01020538 SBS10950.1 HAEF01012610 HAEG01000516 SBR63720.1 GEFH01002336 JAP66245.1 GCES01147828 JAQ38494.1 HAEA01009474 SBQ37954.1 HADZ01000544 SBP64485.1
PZC72163.1 JTDY01002701 KOB70889.1 KQ459838 KPJ19705.1 KQ459460 KPJ00815.1 AGBW02012739 OWR44520.1 KK852539 KDR21781.1 KQ435794 KOX73747.1 GL443836 EFN61587.1 GEDC01004387 JAS32911.1 QOIP01000004 RLU23230.1 KQ978219 KYM96411.1 GL762897 EFZ20569.1 DS235059 EEB11127.1 KQ976500 KYM83261.1 KQ981673 KYN38289.1 ADTU01011209 KK107372 EZA51746.1 KQ980581 KYN15351.1 GL448504 EFN84540.1 GL888932 EGI57265.1 GFDL01010618 JAV24427.1 GFDL01010732 JAV24313.1 KQ983227 KYQ46470.1 GFDL01010933 JAV24112.1 LBMM01001848 KMQ95653.1 GEZM01013114 JAV92742.1 KQ971307 EFA11536.1 JH431613 KQ414775 KOC61307.1 DS233195 EDS28591.1 JXUM01058934 KQ562026 KXJ76872.1 GEZM01013115 JAV92741.1 GDUN01001052 JAN94867.1 NNAY01000821 OXU26335.1 GGMS01014709 MBY83912.1 ABLF02033596 KK854831 PTY19087.1 RQTK01000228 RUS83833.1 LJIG01022709 KRT79118.1 KZ288324 PBC28095.1 NEDP02004420 OWF45695.1 GBHO01022308 GBRD01011615 GBRD01008524 JAG21296.1 JAG54209.1 UFQS01001027 UFQT01001027 SSX08621.1 SSX28537.1 UFQS01000371 UFQT01000371 SSX03260.1 SSX23626.1 GEDC01024887 JAS12411.1 KB203534 ESO84336.1 JH816704 EKC31765.1 GBBI01002821 JAC15891.1 GFTR01007121 JAW09305.1 ACPB03016234 AAGJ04114875 APGK01047232 KB741077 KB631792 ENN74252.1 ERL86121.1 AMQN01001815 KB305766 ELU00735.1 NEVH01023955 PNF17823.1 GDRN01100812 GDRN01100811 JAI58495.1 LJIJ01001303 ODM92184.1 DS469523 EDO46962.1 GANP01002939 JAB81529.1 LSMT01000283 PFX21300.1 AHAT01023625 HAEI01000345 HAEH01007472 SBR71330.1 HAEF01003116 SBR40498.1 HAEG01011855 SBR90830.1 GFAC01002607 JAT96581.1 GEBQ01026606 JAT13371.1 HADY01019312 SBP57797.1 HAEH01020538 SBS10950.1 HAEF01012610 HAEG01000516 SBR63720.1 GEFH01002336 JAP66245.1 GCES01147828 JAQ38494.1 HAEA01009474 SBQ37954.1 HADZ01000544 SBP64485.1
Proteomes
UP000005204
UP000218220
UP000037510
UP000053240
UP000053268
UP000007151
+ More
UP000027135 UP000053105 UP000000311 UP000279307 UP000078542 UP000009046 UP000078540 UP000078541 UP000005205 UP000053097 UP000078492 UP000008237 UP000007755 UP000075809 UP000036403 UP000007266 UP000053825 UP000002320 UP000069940 UP000249989 UP000002358 UP000215335 UP000075900 UP000007819 UP000271974 UP000075885 UP000192223 UP000242457 UP000005203 UP000242188 UP000085678 UP000030746 UP000005408 UP000015103 UP000007110 UP000019118 UP000030742 UP000014760 UP000235965 UP000094527 UP000001593 UP000225706 UP000265120 UP000018468 UP000261540 UP000261660 UP000261600 UP000192224 UP000261500 UP000265020 UP000265140 UP000264800 UP000261440 UP000261400 UP000192220 UP000261560 UP000087266 UP000076420
UP000027135 UP000053105 UP000000311 UP000279307 UP000078542 UP000009046 UP000078540 UP000078541 UP000005205 UP000053097 UP000078492 UP000008237 UP000007755 UP000075809 UP000036403 UP000007266 UP000053825 UP000002320 UP000069940 UP000249989 UP000002358 UP000215335 UP000075900 UP000007819 UP000271974 UP000075885 UP000192223 UP000242457 UP000005203 UP000242188 UP000085678 UP000030746 UP000005408 UP000015103 UP000007110 UP000019118 UP000030742 UP000014760 UP000235965 UP000094527 UP000001593 UP000225706 UP000265120 UP000018468 UP000261540 UP000261660 UP000261600 UP000192224 UP000261500 UP000265020 UP000265140 UP000264800 UP000261440 UP000261400 UP000192220 UP000261560 UP000087266 UP000076420
PRIDE
Interpro
SUPFAM
SSF100920
SSF100920
Gene 3D
ProteinModelPortal
H9J499
A0A2H1WWP8
A0A2A4JV49
A0A2W1BKG2
A0A0L7L6F2
A0A0N1IAN4
+ More
A0A194Q738 A0A212ESQ9 A0A067RD28 A0A0M8ZYX6 E2AYA4 A0A1B6E4R2 A0A3L8DS24 A0A195C8B5 E9IFT0 E0VCM1 A0A195BFA3 A0A195FE38 A0A158NBL2 A0A026W7X6 A0A195DR80 E2BIF2 F4X8J6 A0A1Q3FA67 A0A1Q3F9W8 A0A151WF39 A0A1Q3F997 A0A0J7KZC7 A0A1Y1N4D4 D6W6S7 T1IWF4 A0A0L7QRQ8 B0XHS2 A0A182GEX8 A0A1Y1N4B0 A0A0P6ISR5 K7J104 A0A232F6R2 A0A182RB36 A0A2S2R3J4 J9K9J3 A0A2R7WHG3 A0A3S1HPY0 A0A182P9Y8 A0A0T6AWD1 A0A1W4X400 A0A2A3E8P0 A0A088ALK5 A0A210QAF4 A0A0A9XNT0 A0A336MJK2 A0A336LZW9 A0A1S3J2X2 A0A1B6CG68 V3ZTW0 K1QS76 A0A1S3J3D7 A0A023F3J2 A0A224XAF2 T1HWP1 W4YH44 N6U6Q9 R7U310 A0A2J7PNA7 A0A0P4VZK7 A0A1D2MH41 A7RNP1 V5HT62 A0A2B4RXL3 A0A3P8VA71 W5NJE1 A0A1A8NQL7 A0A3B3SUS2 A0A1A8L7N8 A0A3Q3GN48 A0A3Q3QCX2 A0A3Q3QCV8 A0A1A8QAM4 A0A1E1XBT9 A0A1W5B026 A0A1B6KPM2 A0A1A8ARM9 A0A3B3VBL9 A0A3Q3GG73 A0A1A8RXV6 A0A1A8N3P1 A0A131XJ80 A0A3Q2E0H9 A0A146P3I3 A0A3P8Y692 A0A1A8DYT5 A0A3Q3A706 A0A3B4DNF4 A0A3B5B8J6 A0A2I4CVY7 A0A3B3DWI1 A0A1S3MYN6 A0A2C9LJC6 A0A1A8BDC5
A0A194Q738 A0A212ESQ9 A0A067RD28 A0A0M8ZYX6 E2AYA4 A0A1B6E4R2 A0A3L8DS24 A0A195C8B5 E9IFT0 E0VCM1 A0A195BFA3 A0A195FE38 A0A158NBL2 A0A026W7X6 A0A195DR80 E2BIF2 F4X8J6 A0A1Q3FA67 A0A1Q3F9W8 A0A151WF39 A0A1Q3F997 A0A0J7KZC7 A0A1Y1N4D4 D6W6S7 T1IWF4 A0A0L7QRQ8 B0XHS2 A0A182GEX8 A0A1Y1N4B0 A0A0P6ISR5 K7J104 A0A232F6R2 A0A182RB36 A0A2S2R3J4 J9K9J3 A0A2R7WHG3 A0A3S1HPY0 A0A182P9Y8 A0A0T6AWD1 A0A1W4X400 A0A2A3E8P0 A0A088ALK5 A0A210QAF4 A0A0A9XNT0 A0A336MJK2 A0A336LZW9 A0A1S3J2X2 A0A1B6CG68 V3ZTW0 K1QS76 A0A1S3J3D7 A0A023F3J2 A0A224XAF2 T1HWP1 W4YH44 N6U6Q9 R7U310 A0A2J7PNA7 A0A0P4VZK7 A0A1D2MH41 A7RNP1 V5HT62 A0A2B4RXL3 A0A3P8VA71 W5NJE1 A0A1A8NQL7 A0A3B3SUS2 A0A1A8L7N8 A0A3Q3GN48 A0A3Q3QCX2 A0A3Q3QCV8 A0A1A8QAM4 A0A1E1XBT9 A0A1W5B026 A0A1B6KPM2 A0A1A8ARM9 A0A3B3VBL9 A0A3Q3GG73 A0A1A8RXV6 A0A1A8N3P1 A0A131XJ80 A0A3Q2E0H9 A0A146P3I3 A0A3P8Y692 A0A1A8DYT5 A0A3Q3A706 A0A3B4DNF4 A0A3B5B8J6 A0A2I4CVY7 A0A3B3DWI1 A0A1S3MYN6 A0A2C9LJC6 A0A1A8BDC5
Ontologies
PANTHER
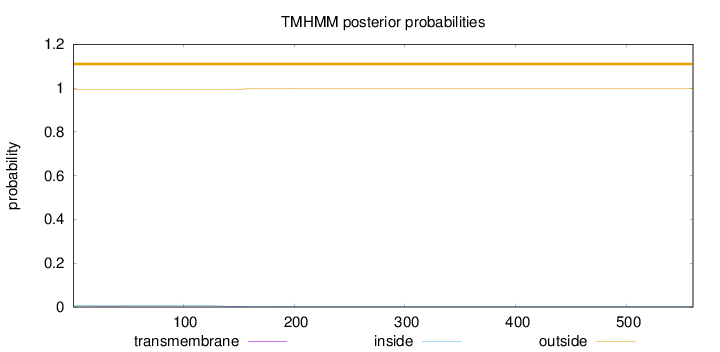
Topology
Length:
560
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0949299999999999
Exp number, first 60 AAs:
0.00742
Total prob of N-in:
0.00573
outside
1 - 560
Population Genetic Test Statistics
Pi
390.375059
Theta
224.954585
Tajima's D
1.876466
CLR
0.607908
CSRT
0.862006899655017
Interpretation
Uncertain
