Pre Gene Modal
BGIBMGA004337
Annotation
PREDICTED:_stromal_membrane-associated_protein_1_[Papilio_machaon]
Location in the cell
Nuclear Reliability : 4.321
Sequence
CDS
ATGACGTCTAAAAGTGAAAAAGATCGTGCTAAACAAATACAGGATCGTTGTCAGAATATATTAGTGCAAATGCTTAAAGATGAAGACAACAAATACTGCGTTGACTGTGATGCAAAAGGTCCACGATGGGCTTCATGGAATTTGGGTATATTCCTGTGCATCCGCTGCGCTGGGATCCATCGGAATCTGGGTGTTCACATCTCCAAAGTGAAGAGTGTCAATCTCGATTCGTGGACTCCAGAGCAAGTGGTGTCGCTACAGCAAATGGGAAACAGCCGCGCCCGGGCCGTGTACGAGGCGAACTTACCAGACTCGTTCAGACGTCCACAAAACGACATGTCCTTGGAATCGTTCATACGCGCCAAATACGAACAGAAGAAATACATTGCGAAGGAGTGGGTGCCCCCGCAGCTGCCCAAAGTCAACTGGGATAAAGAAATAGACGAAGAGATGGATCGACAGAAAAGGAAAAAGAAGTCGGCGACGTCATCGCTCGGACCCCTGCCGGCCCCCGCTGTTGCTGATAAGAAATATAATAAGTCCGACGTGATACCGAGCATACCGAAACCAAAATCTTCAGTCAGTCCTAAGCTCGGTAGGAGCACGCCTCCCGCACACCCGGACACCAGCAAAACGTCCAACGGCTCCGCAGATCTCCTCGGCTTGGACACCGCCAAACCGGACCCCGTGCCCAGCAACGACGACATCTTCTCCAGTTTCTTTTCGGCGCCACAAGAGCGACCAGCCGAGCCGGTCAAGAACGATATCAGCAACGACTTGAAGGTCGAGGAGGAGAACTTCTTCAAGCAACCAGCGCCCTCAGAGAAGGAGAAGTCTAAGCTGACTAAGGACAGCATCTTGGCGTTGTACAGTCAAACGCCGGTGGGAGCTCAGTTTCAGGCGCCGAGCTACCAGTACCCGTCGAATGCTGCTCCGCAAGGGTACTGCGCGATGCCGCAAGCCAGCACCATGCCGTACAACTACCAGCAACCCTTCGCGGCGCCGACCCAGCAACCATTGCAACCACCGTTTGCCAACAGCCAACTCTTCAGCCAACCTCAGCAGACGCAGCCAATCCAACAGCAGTTCGCCAATTTGAACTTGGGACAGGGTTTTCCTAATGCATTCACGCAACAAAACGCGAACTTCGCAAGCAATACATCGTGGCAGTAA
Protein
MTSKSEKDRAKQIQDRCQNILVQMLKDEDNKYCVDCDAKGPRWASWNLGIFLCIRCAGIHRNLGVHISKVKSVNLDSWTPEQVVSLQQMGNSRARAVYEANLPDSFRRPQNDMSLESFIRAKYEQKKYIAKEWVPPQLPKVNWDKEIDEEMDRQKRKKKSATSSLGPLPAPAVADKKYNKSDVIPSIPKPKSSVSPKLGRSTPPAHPDTSKTSNGSADLLGLDTAKPDPVPSNDDIFSSFFSAPQERPAEPVKNDISNDLKVEEENFFKQPAPSEKEKSKLTKDSILALYSQTPVGAQFQAPSYQYPSNAAPQGYCAMPQASTMPYNYQQPFAAPTQQPLQPPFANSQLFSQPQQTQPIQQQFANLNLGQGFPNAFTQQNANFASNTSWQ
Summary
Uniprot
H9J498
A0A2A4JUE2
A0A0N1PHH2
A0A194QBK3
A0A212ESU7
A0A026WY63
+ More
E2B1A8 A0A0J7KIP9 A0A195DXE2 A0A0C9RJN1 A0A232EY52 F4WSR2 K7IQQ6 A0A195FNI9 A0A158NGZ8 A0A151IDE8 A0A151XD91 A0A2J7R0X1 A0A2J7R0X4 A0A2P8XWI5 A0A310SYB7 A0A2J7R0Y2 A0A2J7R0Y1 A0A2W1B9A2 A0A0L7RDL6 A0A2A3EMW7 A0A154PQC9 A0A195B784 A0A0M8ZVN8 A0A088ADM6 A0A1Y1N2Z3 A0A1Y1N2X9 A0A067RKA1 E2C4J0 E9IQH4 A0A0C9QX87 A0A0K8TB35 A0A146M6K8 D6WRH8 A0A1B6MAI2 R4G5J7 A0A0P4VQ62 A0A069DSW3 A0A224XQ24 A0A0V0G6B6 A0A023F8Q6 A0A0K8V9N0 B3MIA7 A0A034W4S9 A0A1A9UVJ7 A0A1B0BX78 B4GCU6 A0A1A9XID8 A0A0L0C5U7 A0A1B6D4X7 A0A1B0AG78 B4KTQ1 Q28Y83 A0A0J9R8E6 A1Z7K6 Q8SWV3 A0A0A1XGJ1 A0A0R3NV94 Q0IG44 A0A0R3NNY7 B4P3E5 A0A0R3NPC5 A0A0P6FM24 A0A1L8DL46 W8BUD6 A0A0P5HCD1 A0A1L8DLG9 E9HY46 A0A1Q3F7I7 A0A0N8DJ12 B3N842 A0A0P5NX43 A0A1I8PB74 A0A1L8DL99 A0A0P5YK14 A0A3B0JMN9 A0A0P4ZPM8 A0A1L8DL71 B0WI04 A0A182GAW2 A0A1Q3F7D5 A0A1Q3F7H9 A0A1W4WF70 A0A0P5UB73 B4LNP1 B4QGN5 T1PDQ7 A0A2S2Q411 A0A0P6HCV2 A0A1E1XK78 A0A0M4EEW0 A0A1W4UJY7 V5I7Y0
E2B1A8 A0A0J7KIP9 A0A195DXE2 A0A0C9RJN1 A0A232EY52 F4WSR2 K7IQQ6 A0A195FNI9 A0A158NGZ8 A0A151IDE8 A0A151XD91 A0A2J7R0X1 A0A2J7R0X4 A0A2P8XWI5 A0A310SYB7 A0A2J7R0Y2 A0A2J7R0Y1 A0A2W1B9A2 A0A0L7RDL6 A0A2A3EMW7 A0A154PQC9 A0A195B784 A0A0M8ZVN8 A0A088ADM6 A0A1Y1N2Z3 A0A1Y1N2X9 A0A067RKA1 E2C4J0 E9IQH4 A0A0C9QX87 A0A0K8TB35 A0A146M6K8 D6WRH8 A0A1B6MAI2 R4G5J7 A0A0P4VQ62 A0A069DSW3 A0A224XQ24 A0A0V0G6B6 A0A023F8Q6 A0A0K8V9N0 B3MIA7 A0A034W4S9 A0A1A9UVJ7 A0A1B0BX78 B4GCU6 A0A1A9XID8 A0A0L0C5U7 A0A1B6D4X7 A0A1B0AG78 B4KTQ1 Q28Y83 A0A0J9R8E6 A1Z7K6 Q8SWV3 A0A0A1XGJ1 A0A0R3NV94 Q0IG44 A0A0R3NNY7 B4P3E5 A0A0R3NPC5 A0A0P6FM24 A0A1L8DL46 W8BUD6 A0A0P5HCD1 A0A1L8DLG9 E9HY46 A0A1Q3F7I7 A0A0N8DJ12 B3N842 A0A0P5NX43 A0A1I8PB74 A0A1L8DL99 A0A0P5YK14 A0A3B0JMN9 A0A0P4ZPM8 A0A1L8DL71 B0WI04 A0A182GAW2 A0A1Q3F7D5 A0A1Q3F7H9 A0A1W4WF70 A0A0P5UB73 B4LNP1 B4QGN5 T1PDQ7 A0A2S2Q411 A0A0P6HCV2 A0A1E1XK78 A0A0M4EEW0 A0A1W4UJY7 V5I7Y0
Pubmed
19121390
26354079
22118469
24508170
20798317
28648823
+ More
21719571 20075255 21347285 29403074 28756777 28004739 24845553 21282665 26823975 18362917 19820115 27129103 26334808 25474469 17994087 25348373 26108605 18057021 15632085 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25830018 17510324 17550304 24495485 21292972 26483478 25315136 29209593
21719571 20075255 21347285 29403074 28756777 28004739 24845553 21282665 26823975 18362917 19820115 27129103 26334808 25474469 17994087 25348373 26108605 18057021 15632085 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25830018 17510324 17550304 24495485 21292972 26483478 25315136 29209593
EMBL
BABH01032619
BABH01032620
NWSH01000561
PCG75645.1
KQ459838
KPJ19704.1
+ More
KQ459460 KPJ00816.1 AGBW02012739 OWR44521.1 KK107063 EZA61010.1 GL444841 EFN60508.1 LBMM01006997 KMQ90127.1 KQ980204 KYN17279.1 GBYB01006923 GBYB01008405 GBYB01008406 JAG76690.1 JAG78172.1 JAG78173.1 NNAY01001691 OXU23209.1 GL888327 EGI62719.1 KQ981387 KYN41991.1 ADTU01015412 ADTU01015413 KQ977971 KYM98412.1 KQ982294 KYQ58323.1 NEVH01008216 PNF34483.1 PNF34484.1 PYGN01001240 PSN36368.1 KQ759820 OAD62744.1 PNF34485.1 PNF34486.1 KZ150212 PZC72162.1 KQ414613 KOC69082.1 KZ288206 PBC33105.1 KQ435037 KZC14125.1 KQ976574 KYM80117.1 KQ435824 KOX72075.1 GEZM01013967 JAV92244.1 GEZM01013968 JAV92243.1 KK852423 KDR24237.1 GL452489 EFN77131.1 GL764790 EFZ17180.1 GBYB01008404 JAG78171.1 GBRD01003210 JAG62611.1 GDHC01004419 JAQ14210.1 KQ971351 EFA06570.2 GEBQ01007034 JAT32943.1 GAHY01000490 JAA77020.1 GDKW01002640 JAI53955.1 GBGD01001706 JAC87183.1 GFTR01005876 JAW10550.1 GECL01002575 JAP03549.1 GBBI01000942 JAC17770.1 GDHF01016791 JAI35523.1 CH902619 EDV35952.1 GAKP01009645 JAC49307.1 JXJN01022118 CH479181 EDW32509.1 JRES01000848 KNC27768.1 GEDC01016595 JAS20703.1 CH933808 EDW09634.1 KRG04778.1 CM000071 EAL26082.2 CM002911 KMY92377.1 AE013599 BT133474 AAM71092.1 AFH97164.1 AY095063 AAM11391.1 GBXI01003788 JAD10504.1 KRT02816.1 CH477274 EAT45167.1 KRT02817.1 CM000157 EDW89418.1 KRT02818.1 GDIQ01046177 JAN48560.1 GFDF01006895 JAV07189.1 GAMC01003738 JAC02818.1 GDIQ01229604 JAK22121.1 GFDF01006894 JAV07190.1 GL733109 EFX63334.1 GFDL01011572 JAV23473.1 GDIP01028951 JAM74764.1 CH954177 EDV59455.1 GDIQ01136485 JAL15241.1 GFDF01006893 JAV07191.1 GDIP01056844 LRGB01000248 JAM46871.1 KZS20017.1 OUUW01000001 SPP73881.1 GDIP01222990 JAJ00412.1 GFDF01006892 JAV07192.1 DS231941 EDS28086.1 JXUM01156941 KQ572609 KXJ68016.1 GFDL01011564 JAV23481.1 GFDL01011582 JAV23463.1 GDIP01119196 JAL84518.1 CH940648 EDW60111.1 CM000362 EDX06260.1 KA646886 AFP61515.1 GGMS01003265 MBY72468.1 GDIQ01021341 JAN73396.1 GFAA01003811 JAT99623.1 CP012524 ALC40635.1 GALX01005650 JAB62816.1
KQ459460 KPJ00816.1 AGBW02012739 OWR44521.1 KK107063 EZA61010.1 GL444841 EFN60508.1 LBMM01006997 KMQ90127.1 KQ980204 KYN17279.1 GBYB01006923 GBYB01008405 GBYB01008406 JAG76690.1 JAG78172.1 JAG78173.1 NNAY01001691 OXU23209.1 GL888327 EGI62719.1 KQ981387 KYN41991.1 ADTU01015412 ADTU01015413 KQ977971 KYM98412.1 KQ982294 KYQ58323.1 NEVH01008216 PNF34483.1 PNF34484.1 PYGN01001240 PSN36368.1 KQ759820 OAD62744.1 PNF34485.1 PNF34486.1 KZ150212 PZC72162.1 KQ414613 KOC69082.1 KZ288206 PBC33105.1 KQ435037 KZC14125.1 KQ976574 KYM80117.1 KQ435824 KOX72075.1 GEZM01013967 JAV92244.1 GEZM01013968 JAV92243.1 KK852423 KDR24237.1 GL452489 EFN77131.1 GL764790 EFZ17180.1 GBYB01008404 JAG78171.1 GBRD01003210 JAG62611.1 GDHC01004419 JAQ14210.1 KQ971351 EFA06570.2 GEBQ01007034 JAT32943.1 GAHY01000490 JAA77020.1 GDKW01002640 JAI53955.1 GBGD01001706 JAC87183.1 GFTR01005876 JAW10550.1 GECL01002575 JAP03549.1 GBBI01000942 JAC17770.1 GDHF01016791 JAI35523.1 CH902619 EDV35952.1 GAKP01009645 JAC49307.1 JXJN01022118 CH479181 EDW32509.1 JRES01000848 KNC27768.1 GEDC01016595 JAS20703.1 CH933808 EDW09634.1 KRG04778.1 CM000071 EAL26082.2 CM002911 KMY92377.1 AE013599 BT133474 AAM71092.1 AFH97164.1 AY095063 AAM11391.1 GBXI01003788 JAD10504.1 KRT02816.1 CH477274 EAT45167.1 KRT02817.1 CM000157 EDW89418.1 KRT02818.1 GDIQ01046177 JAN48560.1 GFDF01006895 JAV07189.1 GAMC01003738 JAC02818.1 GDIQ01229604 JAK22121.1 GFDF01006894 JAV07190.1 GL733109 EFX63334.1 GFDL01011572 JAV23473.1 GDIP01028951 JAM74764.1 CH954177 EDV59455.1 GDIQ01136485 JAL15241.1 GFDF01006893 JAV07191.1 GDIP01056844 LRGB01000248 JAM46871.1 KZS20017.1 OUUW01000001 SPP73881.1 GDIP01222990 JAJ00412.1 GFDF01006892 JAV07192.1 DS231941 EDS28086.1 JXUM01156941 KQ572609 KXJ68016.1 GFDL01011564 JAV23481.1 GFDL01011582 JAV23463.1 GDIP01119196 JAL84518.1 CH940648 EDW60111.1 CM000362 EDX06260.1 KA646886 AFP61515.1 GGMS01003265 MBY72468.1 GDIQ01021341 JAN73396.1 GFAA01003811 JAT99623.1 CP012524 ALC40635.1 GALX01005650 JAB62816.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000007151
UP000053097
+ More
UP000000311 UP000036403 UP000078492 UP000215335 UP000007755 UP000002358 UP000078541 UP000005205 UP000078542 UP000075809 UP000235965 UP000245037 UP000053825 UP000242457 UP000076502 UP000078540 UP000053105 UP000005203 UP000027135 UP000008237 UP000007266 UP000007801 UP000078200 UP000092460 UP000008744 UP000092443 UP000037069 UP000092445 UP000009192 UP000001819 UP000000803 UP000008820 UP000002282 UP000000305 UP000008711 UP000095300 UP000076858 UP000268350 UP000002320 UP000069940 UP000249989 UP000192223 UP000008792 UP000000304 UP000095301 UP000092553 UP000192221
UP000000311 UP000036403 UP000078492 UP000215335 UP000007755 UP000002358 UP000078541 UP000005205 UP000078542 UP000075809 UP000235965 UP000245037 UP000053825 UP000242457 UP000076502 UP000078540 UP000053105 UP000005203 UP000027135 UP000008237 UP000007266 UP000007801 UP000078200 UP000092460 UP000008744 UP000092443 UP000037069 UP000092445 UP000009192 UP000001819 UP000000803 UP000008820 UP000002282 UP000000305 UP000008711 UP000095300 UP000076858 UP000268350 UP000002320 UP000069940 UP000249989 UP000192223 UP000008792 UP000000304 UP000095301 UP000092553 UP000192221
Pfam
PF01412 ArfGap
SUPFAM
SSF57863
SSF57863
Gene 3D
ProteinModelPortal
H9J498
A0A2A4JUE2
A0A0N1PHH2
A0A194QBK3
A0A212ESU7
A0A026WY63
+ More
E2B1A8 A0A0J7KIP9 A0A195DXE2 A0A0C9RJN1 A0A232EY52 F4WSR2 K7IQQ6 A0A195FNI9 A0A158NGZ8 A0A151IDE8 A0A151XD91 A0A2J7R0X1 A0A2J7R0X4 A0A2P8XWI5 A0A310SYB7 A0A2J7R0Y2 A0A2J7R0Y1 A0A2W1B9A2 A0A0L7RDL6 A0A2A3EMW7 A0A154PQC9 A0A195B784 A0A0M8ZVN8 A0A088ADM6 A0A1Y1N2Z3 A0A1Y1N2X9 A0A067RKA1 E2C4J0 E9IQH4 A0A0C9QX87 A0A0K8TB35 A0A146M6K8 D6WRH8 A0A1B6MAI2 R4G5J7 A0A0P4VQ62 A0A069DSW3 A0A224XQ24 A0A0V0G6B6 A0A023F8Q6 A0A0K8V9N0 B3MIA7 A0A034W4S9 A0A1A9UVJ7 A0A1B0BX78 B4GCU6 A0A1A9XID8 A0A0L0C5U7 A0A1B6D4X7 A0A1B0AG78 B4KTQ1 Q28Y83 A0A0J9R8E6 A1Z7K6 Q8SWV3 A0A0A1XGJ1 A0A0R3NV94 Q0IG44 A0A0R3NNY7 B4P3E5 A0A0R3NPC5 A0A0P6FM24 A0A1L8DL46 W8BUD6 A0A0P5HCD1 A0A1L8DLG9 E9HY46 A0A1Q3F7I7 A0A0N8DJ12 B3N842 A0A0P5NX43 A0A1I8PB74 A0A1L8DL99 A0A0P5YK14 A0A3B0JMN9 A0A0P4ZPM8 A0A1L8DL71 B0WI04 A0A182GAW2 A0A1Q3F7D5 A0A1Q3F7H9 A0A1W4WF70 A0A0P5UB73 B4LNP1 B4QGN5 T1PDQ7 A0A2S2Q411 A0A0P6HCV2 A0A1E1XK78 A0A0M4EEW0 A0A1W4UJY7 V5I7Y0
E2B1A8 A0A0J7KIP9 A0A195DXE2 A0A0C9RJN1 A0A232EY52 F4WSR2 K7IQQ6 A0A195FNI9 A0A158NGZ8 A0A151IDE8 A0A151XD91 A0A2J7R0X1 A0A2J7R0X4 A0A2P8XWI5 A0A310SYB7 A0A2J7R0Y2 A0A2J7R0Y1 A0A2W1B9A2 A0A0L7RDL6 A0A2A3EMW7 A0A154PQC9 A0A195B784 A0A0M8ZVN8 A0A088ADM6 A0A1Y1N2Z3 A0A1Y1N2X9 A0A067RKA1 E2C4J0 E9IQH4 A0A0C9QX87 A0A0K8TB35 A0A146M6K8 D6WRH8 A0A1B6MAI2 R4G5J7 A0A0P4VQ62 A0A069DSW3 A0A224XQ24 A0A0V0G6B6 A0A023F8Q6 A0A0K8V9N0 B3MIA7 A0A034W4S9 A0A1A9UVJ7 A0A1B0BX78 B4GCU6 A0A1A9XID8 A0A0L0C5U7 A0A1B6D4X7 A0A1B0AG78 B4KTQ1 Q28Y83 A0A0J9R8E6 A1Z7K6 Q8SWV3 A0A0A1XGJ1 A0A0R3NV94 Q0IG44 A0A0R3NNY7 B4P3E5 A0A0R3NPC5 A0A0P6FM24 A0A1L8DL46 W8BUD6 A0A0P5HCD1 A0A1L8DLG9 E9HY46 A0A1Q3F7I7 A0A0N8DJ12 B3N842 A0A0P5NX43 A0A1I8PB74 A0A1L8DL99 A0A0P5YK14 A0A3B0JMN9 A0A0P4ZPM8 A0A1L8DL71 B0WI04 A0A182GAW2 A0A1Q3F7D5 A0A1Q3F7H9 A0A1W4WF70 A0A0P5UB73 B4LNP1 B4QGN5 T1PDQ7 A0A2S2Q411 A0A0P6HCV2 A0A1E1XK78 A0A0M4EEW0 A0A1W4UJY7 V5I7Y0
PDB
2IQJ
E-value=1.83155e-45,
Score=460
Ontologies
GO
Topology
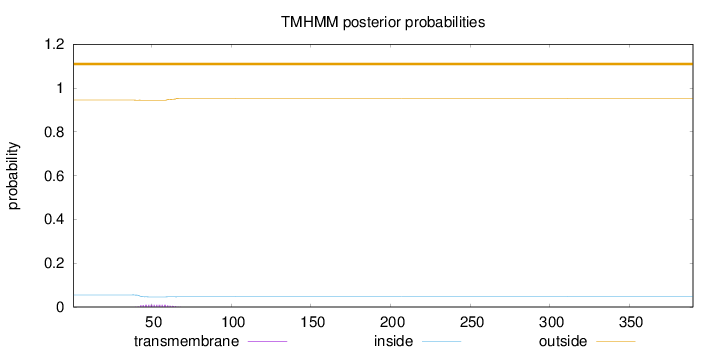
Length:
390
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.19766
Exp number, first 60 AAs:
0.17064
Total prob of N-in:
0.05517
outside
1 - 390
Population Genetic Test Statistics
Pi
177.325828
Theta
133.856147
Tajima's D
0.510099
CLR
0.000402
CSRT
0.520073996300185
Interpretation
Uncertain
