Gene
KWMTBOMO12087
Pre Gene Modal
BGIBMGA004279
Annotation
PREDICTED:_phosphoinositide_3-kinase_regulatory_subunit_4_isoform_X2_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.523
Sequence
CDS
ATGGGCCACAGCGCTAGTTACGTGCAGTATTGTATGGCGGCTTGCAGAGTTGAACTGAGAGATTTAACCGCTCGGATGCAGCAAAAATTTCAAAAGTCTTTGAGGAGTCATGAGAACGCAGCAGAGAGCAGCGACGACATAATACCGCCTGCGTCGTGGCGTCCAGGCAACCAGCTGTTGGCTTATCTACACGAACACAAGGCCCAGGTCAACGACCTGGTGACGCTACCCGCGCACCGGAACATGTTCGGCTCGTGCTCCAACGACGGCACCGTCCGGCTGTGGGACGTGGCCAAGCTGCAGGGGCACGCGCACATCAACAGATCGAAGTCGATGTACAACAGGAGCGCGGGCCCGGTGATATCGTTGGCTTCGTGCGAGGCGGGGCAGTCTTTCGCGGCCGCCACCCAGGACGGATCCGTGTTCGTGCTCAGATTAGACAGCGCGTCGTCCCGCATGACCCTCTCGCAGTGCCGTCAGCTTGAGGTCGGGGCGGAGGGGGAGTGCTGCGCCATGACCTGCGCCGGCGGAGTCATATCGTACGCGACCCTCCGCTCGAAGATCGTCGGTTGGGACCTAAGGACACCTGGTAACGCGTGGAAACTGGACGGAGATCTGAAGCAGGGCGTGATAACGTGCCTGTACGCCTCCAGCAGCGGGTACGTAGTGGCGGGCACCTCCAGCGGCGTCCTCAGCGTCTGGGACCTGCGCTTCCAGCTGCCCATCACCACGCTCAAGCATCCCAACGCGGAGCGGGTGCGGCGCGTGCACGGCGCCCCGGGCTCGTCGCTGGTGTGGGCGGCGGGCGCTCGCGACGCGGCCGCCAGCTGCCTGGAGACCACGCAGCGCACGCACGCACTCTGGACCTGCACCAGCTCGCCACAGCCGCTGCACTACGCCGCACCCGCGTCCCAGTACATCAGCGCGATGCTGTGTGACGCTGAAGGTCGGTTCGTGATAACGGGCGGCTCGGACCTCCGGCTGCGCTACTGGGACCTGCAGCATCCCGAAGACTCGTACATCCTGCTGCACGCGCCGTCTGATCATCTCAAACACTCACCTAATTCGGTCAAATACAGGAGTCGCATTGTAGAGGGCACCACCGTCATACAGGAGTGTTGCAAGCTGAACCCCAACTCGCCACCCCCCGCACTGGAGGACAACGTGTACCGTACTGTCGAGTCTAGGTCGTACCACCACACGGCGCCCATCACGGACCTCACCCTGGTCGAGTCCAACAAACACTACCTGGTCAGTTCCTCCGCTGACGGCGTTATCAATGTTTGGAAGTAA
Protein
MGHSASYVQYCMAACRVELRDLTARMQQKFQKSLRSHENAAESSDDIIPPASWRPGNQLLAYLHEHKAQVNDLVTLPAHRNMFGSCSNDGTVRLWDVAKLQGHAHINRSKSMYNRSAGPVISLASCEAGQSFAAATQDGSVFVLRLDSASSRMTLSQCRQLEVGAEGECCAMTCAGGVISYATLRSKIVGWDLRTPGNAWKLDGDLKQGVITCLYASSSGYVVAGTSSGVLSVWDLRFQLPITTLKHPNAERVRRVHGAPGSSLVWAAGARDAAASCLETTQRTHALWTCTSSPQPLHYAAPASQYISAMLCDAEGRFVITGGSDLRLRYWDLQHPEDSYILLHAPSDHLKHSPNSVKYRSRIVEGTTVIQECCKLNPNSPPPALEDNVYRTVESRSYHHTAPITDLTLVESNKHYLVSSSADGVINVWK
Summary
Uniprot
H9J440
A0A194Q5J6
A0A2H1VFW9
A0A212ESQ2
A0A0N1IGU5
A0A2J7QKL4
+ More
E9IQS5 A0A0T6AWP7 A0A3L8DXR8 A0A026WRN9 A0A0C9R475 E2B139 A0A067QVH4 A0A1W4X9V5 A0A195CP48 A0A1Y1L7L3 D6X2A3 A0A195E689 R7TU23 F4WP24 A0A158NGB3 A0A151WM29 A0A195BA35 K7JA52 A0A2S2NV17 A0A1J1HLS6 A0A2S2R9T1 A0A195EV18 A0A2A3E980 J9JZ69 A0A232EL23 A0A2H8TZ70 A0A088ABY1 A0A1L8DLV4 A0A0L7RBS4 A0A1B6KTS4 A0A1B6LYU7 A0A154PGF4 A0A1B0DMA7 A0A0A9XEH8 A0A336MEQ7 A0A0N0U3V0 A0A3R7SZY0 A0A146L0G8 A0A0L8GE65 A0A1S3IHZ3 E0VUY6 E2BB70 A0A1Q3FY40 C3XYM7 A0A0P4VQM1 C3ZYW7 A0A224YVU7 V5H528 A0A1S4F3D8 Q17GU5 V5HA25 A0A182GGB0 A0A182GFJ8 A0A224XIF7 A0A2R7VVD6 A0A131YJM6 A0A131YHQ0 A0A1I8MHQ4 A0A034VP91 A0A034VMM2 A0A034VNK6 T1I0F9 A0A224YVV4 A0A2R5LC50 A0A3Q3FL54 W8B2U2 A0A146Y7A1 A0A3Q3FLN7 A0A0A1WIG6 A0A023F3C6 A0A1A8KI66 A0A1A8V619 A0A1B6DEI4 A0A1A8ALV8 A0A1A8BI21 L7M5W0 A0A1A8P3V3 A0A1A8RFE7 A0A1A8PKM4 A0A1A8J6P7 A0A1A7YFJ7 A0A1I8PYQ0 A0A3Q2E000 A0A3Q1FQK3 A0A1A8GG30 A0A1W5AGW7 A0A2D0RT31 A0A2D0RS36 A0A3Q2R2F5
E9IQS5 A0A0T6AWP7 A0A3L8DXR8 A0A026WRN9 A0A0C9R475 E2B139 A0A067QVH4 A0A1W4X9V5 A0A195CP48 A0A1Y1L7L3 D6X2A3 A0A195E689 R7TU23 F4WP24 A0A158NGB3 A0A151WM29 A0A195BA35 K7JA52 A0A2S2NV17 A0A1J1HLS6 A0A2S2R9T1 A0A195EV18 A0A2A3E980 J9JZ69 A0A232EL23 A0A2H8TZ70 A0A088ABY1 A0A1L8DLV4 A0A0L7RBS4 A0A1B6KTS4 A0A1B6LYU7 A0A154PGF4 A0A1B0DMA7 A0A0A9XEH8 A0A336MEQ7 A0A0N0U3V0 A0A3R7SZY0 A0A146L0G8 A0A0L8GE65 A0A1S3IHZ3 E0VUY6 E2BB70 A0A1Q3FY40 C3XYM7 A0A0P4VQM1 C3ZYW7 A0A224YVU7 V5H528 A0A1S4F3D8 Q17GU5 V5HA25 A0A182GGB0 A0A182GFJ8 A0A224XIF7 A0A2R7VVD6 A0A131YJM6 A0A131YHQ0 A0A1I8MHQ4 A0A034VP91 A0A034VMM2 A0A034VNK6 T1I0F9 A0A224YVV4 A0A2R5LC50 A0A3Q3FL54 W8B2U2 A0A146Y7A1 A0A3Q3FLN7 A0A0A1WIG6 A0A023F3C6 A0A1A8KI66 A0A1A8V619 A0A1B6DEI4 A0A1A8ALV8 A0A1A8BI21 L7M5W0 A0A1A8P3V3 A0A1A8RFE7 A0A1A8PKM4 A0A1A8J6P7 A0A1A7YFJ7 A0A1I8PYQ0 A0A3Q2E000 A0A3Q1FQK3 A0A1A8GG30 A0A1W5AGW7 A0A2D0RT31 A0A2D0RS36 A0A3Q2R2F5
Pubmed
EMBL
BABH01032618
BABH01032619
KQ459460
KPJ00818.1
ODYU01002346
SOQ39725.1
+ More
AGBW02012739 OWR44522.1 KQ459838 KPJ19703.1 NEVH01013262 PNF29121.1 GL764900 EFZ17067.1 LJIG01022667 KRT79355.1 QOIP01000003 RLU24739.1 KK107139 EZA57769.1 GBYB01002700 JAG72467.1 GL444771 EFN60612.1 KK852902 KDR14050.1 KQ977574 KYN01884.1 GEZM01068267 JAV67057.1 KQ971371 EFA10236.2 KQ979608 KYN20364.1 AMQN01010912 KB308566 ELT97398.1 GL888243 EGI63955.1 ADTU01015125 KQ982944 KYQ48906.1 KQ976542 KYM81059.1 AAZX01001625 GGMR01008358 MBY20977.1 CVRI01000010 CRK88989.1 GGMS01017227 MBY86430.1 KQ981965 KYN31737.1 KZ288338 PBC27832.1 ABLF02009436 ABLF02009437 NNAY01003660 OXU19045.1 GFXV01007394 MBW19199.1 GFDF01006693 JAV07391.1 KQ414617 KOC68309.1 GEBQ01025125 JAT14852.1 GEBQ01011177 JAT28800.1 KQ434899 KZC10923.1 AJVK01016709 GBHO01026401 GBRD01017427 JAG17203.1 JAG48400.1 UFQS01001084 UFQT01001084 SSX08903.1 SSX28814.1 KQ435851 KOX70771.1 QCYY01000677 ROT83496.1 GDHC01017763 JAQ00866.1 KQ422315 KOF75124.1 DS235797 EEB17192.1 GL446946 EFN87043.1 GFDL01002571 JAV32474.1 GG666473 EEN66912.1 GDRN01106043 GDRN01106042 JAI57638.1 GG666738 EEN42238.1 GFPF01010591 MAA21737.1 GANP01015066 JAB69402.1 CH477256 EAT45842.1 GANP01004278 JAB80190.1 JXUM01001496 KQ560122 KXJ84353.1 JXUM01059703 KQ562068 KXJ76749.1 GFTR01008503 JAW07923.1 KK854110 PTY11463.1 GEDV01009789 JAP78768.1 GEDV01009788 JAP78769.1 GAKP01015579 JAC43373.1 GAKP01015580 JAC43372.1 GAKP01015582 JAC43370.1 ACPB03006600 GFPF01010590 MAA21736.1 GGLE01002978 MBY07104.1 GAMC01018994 JAB87561.1 GCES01036891 JAR49432.1 GBXI01016012 GBXI01000414 JAC98279.1 JAD13878.1 GBBI01002991 JAC15721.1 HAEE01011339 SBR31389.1 HAEJ01014794 SBS55251.1 GEDC01013242 JAS24056.1 HADY01017519 SBP56004.1 HADZ01002597 HAEA01015897 SBP66538.1 GACK01005363 JAA59671.1 HAEI01001325 SBR75692.1 HAEH01016172 SBS04108.1 HAEF01008966 HAEG01008319 SBR81562.1 HAED01018726 SBR05171.1 HADW01002218 HADX01006478 SBP28710.1 HAEB01010796 HAEC01002648 SBQ70725.1
AGBW02012739 OWR44522.1 KQ459838 KPJ19703.1 NEVH01013262 PNF29121.1 GL764900 EFZ17067.1 LJIG01022667 KRT79355.1 QOIP01000003 RLU24739.1 KK107139 EZA57769.1 GBYB01002700 JAG72467.1 GL444771 EFN60612.1 KK852902 KDR14050.1 KQ977574 KYN01884.1 GEZM01068267 JAV67057.1 KQ971371 EFA10236.2 KQ979608 KYN20364.1 AMQN01010912 KB308566 ELT97398.1 GL888243 EGI63955.1 ADTU01015125 KQ982944 KYQ48906.1 KQ976542 KYM81059.1 AAZX01001625 GGMR01008358 MBY20977.1 CVRI01000010 CRK88989.1 GGMS01017227 MBY86430.1 KQ981965 KYN31737.1 KZ288338 PBC27832.1 ABLF02009436 ABLF02009437 NNAY01003660 OXU19045.1 GFXV01007394 MBW19199.1 GFDF01006693 JAV07391.1 KQ414617 KOC68309.1 GEBQ01025125 JAT14852.1 GEBQ01011177 JAT28800.1 KQ434899 KZC10923.1 AJVK01016709 GBHO01026401 GBRD01017427 JAG17203.1 JAG48400.1 UFQS01001084 UFQT01001084 SSX08903.1 SSX28814.1 KQ435851 KOX70771.1 QCYY01000677 ROT83496.1 GDHC01017763 JAQ00866.1 KQ422315 KOF75124.1 DS235797 EEB17192.1 GL446946 EFN87043.1 GFDL01002571 JAV32474.1 GG666473 EEN66912.1 GDRN01106043 GDRN01106042 JAI57638.1 GG666738 EEN42238.1 GFPF01010591 MAA21737.1 GANP01015066 JAB69402.1 CH477256 EAT45842.1 GANP01004278 JAB80190.1 JXUM01001496 KQ560122 KXJ84353.1 JXUM01059703 KQ562068 KXJ76749.1 GFTR01008503 JAW07923.1 KK854110 PTY11463.1 GEDV01009789 JAP78768.1 GEDV01009788 JAP78769.1 GAKP01015579 JAC43373.1 GAKP01015580 JAC43372.1 GAKP01015582 JAC43370.1 ACPB03006600 GFPF01010590 MAA21736.1 GGLE01002978 MBY07104.1 GAMC01018994 JAB87561.1 GCES01036891 JAR49432.1 GBXI01016012 GBXI01000414 JAC98279.1 JAD13878.1 GBBI01002991 JAC15721.1 HAEE01011339 SBR31389.1 HAEJ01014794 SBS55251.1 GEDC01013242 JAS24056.1 HADY01017519 SBP56004.1 HADZ01002597 HAEA01015897 SBP66538.1 GACK01005363 JAA59671.1 HAEI01001325 SBR75692.1 HAEH01016172 SBS04108.1 HAEF01008966 HAEG01008319 SBR81562.1 HAED01018726 SBR05171.1 HADW01002218 HADX01006478 SBP28710.1 HAEB01010796 HAEC01002648 SBQ70725.1
Proteomes
UP000005204
UP000053268
UP000007151
UP000053240
UP000235965
UP000279307
+ More
UP000053097 UP000000311 UP000027135 UP000192223 UP000078542 UP000007266 UP000078492 UP000014760 UP000007755 UP000005205 UP000075809 UP000078540 UP000002358 UP000183832 UP000078541 UP000242457 UP000007819 UP000215335 UP000005203 UP000053825 UP000076502 UP000092462 UP000053105 UP000283509 UP000053454 UP000085678 UP000009046 UP000008237 UP000001554 UP000008820 UP000069940 UP000249989 UP000095301 UP000015103 UP000261660 UP000095300 UP000265020 UP000257200 UP000192224 UP000221080 UP000265000
UP000053097 UP000000311 UP000027135 UP000192223 UP000078542 UP000007266 UP000078492 UP000014760 UP000007755 UP000005205 UP000075809 UP000078540 UP000002358 UP000183832 UP000078541 UP000242457 UP000007819 UP000215335 UP000005203 UP000053825 UP000076502 UP000092462 UP000053105 UP000283509 UP000053454 UP000085678 UP000009046 UP000008237 UP000001554 UP000008820 UP000069940 UP000249989 UP000095301 UP000015103 UP000261660 UP000095300 UP000265020 UP000257200 UP000192224 UP000221080 UP000265000
PRIDE
Interpro
IPR015943
WD40/YVTN_repeat-like_dom_sf
+ More
IPR000719 Prot_kinase_dom
IPR017986 WD40_repeat_dom
IPR001680 WD40_repeat
IPR016024 ARM-type_fold
IPR011989 ARM-like
IPR011009 Kinase-like_dom_sf
IPR008271 Ser/Thr_kinase_AS
IPR036322 WD40_repeat_dom_sf
IPR020472 G-protein_beta_WD-40_rep
IPR019775 WD40_repeat_CS
IPR021133 HEAT_type_2
IPR000357 HEAT
IPR000719 Prot_kinase_dom
IPR017986 WD40_repeat_dom
IPR001680 WD40_repeat
IPR016024 ARM-type_fold
IPR011989 ARM-like
IPR011009 Kinase-like_dom_sf
IPR008271 Ser/Thr_kinase_AS
IPR036322 WD40_repeat_dom_sf
IPR020472 G-protein_beta_WD-40_rep
IPR019775 WD40_repeat_CS
IPR021133 HEAT_type_2
IPR000357 HEAT
Gene 3D
ProteinModelPortal
H9J440
A0A194Q5J6
A0A2H1VFW9
A0A212ESQ2
A0A0N1IGU5
A0A2J7QKL4
+ More
E9IQS5 A0A0T6AWP7 A0A3L8DXR8 A0A026WRN9 A0A0C9R475 E2B139 A0A067QVH4 A0A1W4X9V5 A0A195CP48 A0A1Y1L7L3 D6X2A3 A0A195E689 R7TU23 F4WP24 A0A158NGB3 A0A151WM29 A0A195BA35 K7JA52 A0A2S2NV17 A0A1J1HLS6 A0A2S2R9T1 A0A195EV18 A0A2A3E980 J9JZ69 A0A232EL23 A0A2H8TZ70 A0A088ABY1 A0A1L8DLV4 A0A0L7RBS4 A0A1B6KTS4 A0A1B6LYU7 A0A154PGF4 A0A1B0DMA7 A0A0A9XEH8 A0A336MEQ7 A0A0N0U3V0 A0A3R7SZY0 A0A146L0G8 A0A0L8GE65 A0A1S3IHZ3 E0VUY6 E2BB70 A0A1Q3FY40 C3XYM7 A0A0P4VQM1 C3ZYW7 A0A224YVU7 V5H528 A0A1S4F3D8 Q17GU5 V5HA25 A0A182GGB0 A0A182GFJ8 A0A224XIF7 A0A2R7VVD6 A0A131YJM6 A0A131YHQ0 A0A1I8MHQ4 A0A034VP91 A0A034VMM2 A0A034VNK6 T1I0F9 A0A224YVV4 A0A2R5LC50 A0A3Q3FL54 W8B2U2 A0A146Y7A1 A0A3Q3FLN7 A0A0A1WIG6 A0A023F3C6 A0A1A8KI66 A0A1A8V619 A0A1B6DEI4 A0A1A8ALV8 A0A1A8BI21 L7M5W0 A0A1A8P3V3 A0A1A8RFE7 A0A1A8PKM4 A0A1A8J6P7 A0A1A7YFJ7 A0A1I8PYQ0 A0A3Q2E000 A0A3Q1FQK3 A0A1A8GG30 A0A1W5AGW7 A0A2D0RT31 A0A2D0RS36 A0A3Q2R2F5
E9IQS5 A0A0T6AWP7 A0A3L8DXR8 A0A026WRN9 A0A0C9R475 E2B139 A0A067QVH4 A0A1W4X9V5 A0A195CP48 A0A1Y1L7L3 D6X2A3 A0A195E689 R7TU23 F4WP24 A0A158NGB3 A0A151WM29 A0A195BA35 K7JA52 A0A2S2NV17 A0A1J1HLS6 A0A2S2R9T1 A0A195EV18 A0A2A3E980 J9JZ69 A0A232EL23 A0A2H8TZ70 A0A088ABY1 A0A1L8DLV4 A0A0L7RBS4 A0A1B6KTS4 A0A1B6LYU7 A0A154PGF4 A0A1B0DMA7 A0A0A9XEH8 A0A336MEQ7 A0A0N0U3V0 A0A3R7SZY0 A0A146L0G8 A0A0L8GE65 A0A1S3IHZ3 E0VUY6 E2BB70 A0A1Q3FY40 C3XYM7 A0A0P4VQM1 C3ZYW7 A0A224YVU7 V5H528 A0A1S4F3D8 Q17GU5 V5HA25 A0A182GGB0 A0A182GFJ8 A0A224XIF7 A0A2R7VVD6 A0A131YJM6 A0A131YHQ0 A0A1I8MHQ4 A0A034VP91 A0A034VMM2 A0A034VNK6 T1I0F9 A0A224YVV4 A0A2R5LC50 A0A3Q3FL54 W8B2U2 A0A146Y7A1 A0A3Q3FLN7 A0A0A1WIG6 A0A023F3C6 A0A1A8KI66 A0A1A8V619 A0A1B6DEI4 A0A1A8ALV8 A0A1A8BI21 L7M5W0 A0A1A8P3V3 A0A1A8RFE7 A0A1A8PKM4 A0A1A8J6P7 A0A1A7YFJ7 A0A1I8PYQ0 A0A3Q2E000 A0A3Q1FQK3 A0A1A8GG30 A0A1W5AGW7 A0A2D0RT31 A0A2D0RS36 A0A3Q2R2F5
PDB
4LG8
E-value=2.3544e-07,
Score=132
Ontologies
GO
Topology
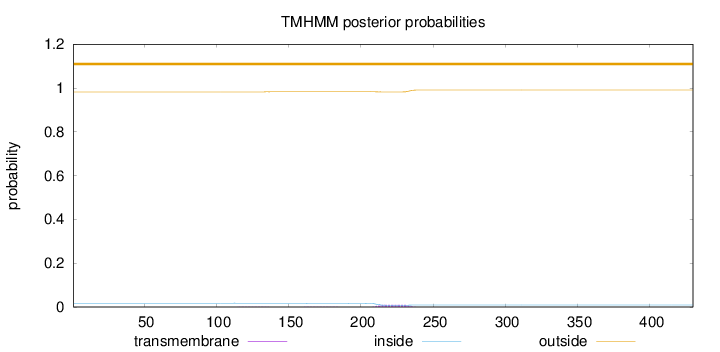
Length:
430
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.24551
Exp number, first 60 AAs:
0.00051
Total prob of N-in:
0.01753
outside
1 - 430
Population Genetic Test Statistics
Pi
207.321291
Theta
167.499844
Tajima's D
1.063979
CLR
0.108069
CSRT
0.676266186690666
Interpretation
Uncertain
