Gene
KWMTBOMO12085
Pre Gene Modal
BGIBMGA004280
Annotation
PREDICTED:_glutamine_synthetase_2_cytoplasmic_[Papilio_machaon]
Full name
Glutamine synthetase
+ More
Glutamine synthetase 2 cytoplasmic
Glutamine synthetase 2 cytoplasmic
Alternative Name
Glutamate--ammonia ligase 2
Location in the cell
Cytoplasmic Reliability : 1.954 Nuclear Reliability : 1.453
Sequence
CDS
ATGTCGGAAACTCAAAACAAAGTGCAAGATAATCCAAAAATTTTATCGGGTCCCGTGCTACAGAACTCTCCGAATGCATTCCTCTCGAAGACACTGTTGGACCGGTATAGAGATCTACCGCTGCCGGAAGACAAGGTCCTCGCCACATACGTATGGATCGACGGCTCCGGCGAGCATGTTCGTTGCAAAGACAGGACTTTGAACTACATTCCAGAAACTCCTAAAGACCTGCCGATATGGAATTACGACGGCAGCTCTACAGGCCAGACGACAAGCACGAACTCAGACACGCACCTGAAGCCCCAGGCCATATACAGGGACCCGTTCCGGCGCGGAAACCACGTGCTTGTCATGTGCGACACATACGATTACAGCATGCAACCAACAAAGACAAACCACCGTATTAAATGCCAAGAAGCGTACGACCGCTGCAGAGACCACGAGCCGTGGTTCGGCATAGAGCAAGAGTACGTGCTCCTCGACTCTGACCTTAGACCGTTTGGGTGGCCTACAACTGGAAGCCCTCCTGCCCAGGGACCATATTATTGCGGCGTTGGAGCTAACAAGGTCTTCGCGAGGGATCTCATTGAAGCCCATTATAAATGCTGTCTGTATGCTGGAGTGCCGATCACAGGTACGAACGCTGAGGCGATGCTCTCGCAGTGGGAGTTCCAAGTCGGGCCCTCCGTTGGCGTGTCAGCTGGAGATGACCTGTGGATAGCTCGTTTTATCCTGCATCGTCTGGCTGAAGAGTTCGGAGTCATCGTCACTTTCGACCCCAAACCGGTGTCGGATTGGAATGGTTCGGGTGCTCACCACAATTTCTCGACAAAGAAAATGCGAGATGAAGGGGGAATTACTGAAATCGAGAGGGCGATTGAAAAGCTCTCCAAAGTGCACATGAAGCACATTAAGGTATACGACCCTCGCGGCGGCAAGGATAACGAGCGCCGACTGACAGGACTCCACGAAACCGCTAGCATTCATGATTTTAGTTCGGGTGTTGCTAGCCGCAGTGCCAGTATAAGAATACCTCGGCAGGTGGAGGCAGAAAAACGCGGGTACCTCGAGGATCGGCGCCCGGCTTCGAACTGCGATCCGTACTCTGTGACCGATGCACTCATGCGCACTTGTATACTTGACGAATTATAA
Protein
MSETQNKVQDNPKILSGPVLQNSPNAFLSKTLLDRYRDLPLPEDKVLATYVWIDGSGEHVRCKDRTLNYIPETPKDLPIWNYDGSSTGQTTSTNSDTHLKPQAIYRDPFRRGNHVLVMCDTYDYSMQPTKTNHRIKCQEAYDRCRDHEPWFGIEQEYVLLDSDLRPFGWPTTGSPPAQGPYYCGVGANKVFARDLIEAHYKCCLYAGVPITGTNAEAMLSQWEFQVGPSVGVSAGDDLWIARFILHRLAEEFGVIVTFDPKPVSDWNGSGAHHNFSTKKMRDEGGITEIERAIEKLSKVHMKHIKVYDPRGGKDNERRLTGLHETASIHDFSSGVASRSASIRIPRQVEAEKRGYLEDRRPASNCDPYSVTDALMRTCILDEL
Summary
Catalytic Activity
ATP + L-glutamate + NH4(+) = ADP + H(+) + L-glutamine + phosphate
Subunit
Homooctamer.
Similarity
Belongs to the glutamine synthetase family.
Keywords
Alternative splicing
ATP-binding
Complete proteome
Cytoplasm
Ligase
Nucleotide-binding
Reference proteome
Feature
chain Glutamine synthetase 2 cytoplasmic
splice variant In isoform A.
splice variant In isoform A.
Uniprot
H9J441
A0A2A4J4U6
A0A2W1BMY2
I4DJ32
A0A0G3VIA2
S4PCI7
+ More
H9ITV0 A0A194QUW7 A0A212ETE4 A0A2J7RJB5 D6WFH1 A0A067R5C5 D3TPP6 V5G3M6 A0A1A9UHR5 A0A0M9A1M4 A0A0M5J396 A0A1B0B749 A0A1B0AIW9 A0A068TK10 A0A1A9Y1D7 A0A1B6IB03 J3JTU0 A0A1J1HER3 A0A1Y1KMB3 A0A1I9WLM6 A0A0A1XT45 W8BUA3 A0A0A1WVL5 A0A0K8VEK6 A0A034VVT9 A0A0K8VUW5 A0A034VRN0 A0A0L0C841 A0A1B6IKT6 A0A2L1DH34 R4G5A5 A0A1I8NDX3 A0A0P4VPJ5 M9V250 A0A0L7R3F9 A0A1W4V0I0 A0A1B6EY34 A0A195BGP1 Q16J32 A0A1B6FLE3 Q2KQ97 S5NFE0 A0A1W4UNL4 A0A195C1J9 A0A0A9WUK9 A0A151XJR8 B4N1T9 A0A1Q3G582 B4L4A8 B4MCX6 A0A3B0JXI0 V9IDX6 A0A0Q9VZ95 A0A2A3E9Y7 B4JIW8 A0A088A2A8 A0A0Q9WMW7 A0A194PS35 E2BUV7 A0A1Q3G4Z2 A0A151JWT9 D0QWE3 A0A026W7I7 Q29H28 B4H0G6 B4PYB1 X2JJG8 P20478 C8VV58 X2JDA5 A0A1L8E2I2 A0A1B6CN61 F4WK81 A0A336LVS2 B3NUD5 P20478-3 Q494K4 A0A0R1EIC3 B4IK13 A0A0G2QRP6 A0A3B0JXJ2 A0A182U6F2 A0A182XIC8 A0A182LFG0 Q7QBX9 A0A084VJR5 G9C5D9 A0A0V0G2W8 E2ATW3 A0A182V4E1 F5HK76 A0A182HTB3 A0A023EQU3 A0A0Q5T3A2
H9ITV0 A0A194QUW7 A0A212ETE4 A0A2J7RJB5 D6WFH1 A0A067R5C5 D3TPP6 V5G3M6 A0A1A9UHR5 A0A0M9A1M4 A0A0M5J396 A0A1B0B749 A0A1B0AIW9 A0A068TK10 A0A1A9Y1D7 A0A1B6IB03 J3JTU0 A0A1J1HER3 A0A1Y1KMB3 A0A1I9WLM6 A0A0A1XT45 W8BUA3 A0A0A1WVL5 A0A0K8VEK6 A0A034VVT9 A0A0K8VUW5 A0A034VRN0 A0A0L0C841 A0A1B6IKT6 A0A2L1DH34 R4G5A5 A0A1I8NDX3 A0A0P4VPJ5 M9V250 A0A0L7R3F9 A0A1W4V0I0 A0A1B6EY34 A0A195BGP1 Q16J32 A0A1B6FLE3 Q2KQ97 S5NFE0 A0A1W4UNL4 A0A195C1J9 A0A0A9WUK9 A0A151XJR8 B4N1T9 A0A1Q3G582 B4L4A8 B4MCX6 A0A3B0JXI0 V9IDX6 A0A0Q9VZ95 A0A2A3E9Y7 B4JIW8 A0A088A2A8 A0A0Q9WMW7 A0A194PS35 E2BUV7 A0A1Q3G4Z2 A0A151JWT9 D0QWE3 A0A026W7I7 Q29H28 B4H0G6 B4PYB1 X2JJG8 P20478 C8VV58 X2JDA5 A0A1L8E2I2 A0A1B6CN61 F4WK81 A0A336LVS2 B3NUD5 P20478-3 Q494K4 A0A0R1EIC3 B4IK13 A0A0G2QRP6 A0A3B0JXJ2 A0A182U6F2 A0A182XIC8 A0A182LFG0 Q7QBX9 A0A084VJR5 G9C5D9 A0A0V0G2W8 E2ATW3 A0A182V4E1 F5HK76 A0A182HTB3 A0A023EQU3 A0A0Q5T3A2
EC Number
6.3.1.2
Pubmed
19121390
28756777
22651552
23622113
26354079
22118469
+ More
18362917 19820115 24845553 20353571 29403074 22516182 28004739 27538518 25830018 24495485 25348373 26108605 25315136 27129103 17510324 15804581 23791183 25401762 17994087 20798317 19859648 24508170 15632085 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 1969491 12537569 21719571 20966253 12364791 14747013 17210077 24438588 21835214 24945155
18362917 19820115 24845553 20353571 29403074 22516182 28004739 27538518 25830018 24495485 25348373 26108605 25315136 27129103 17510324 15804581 23791183 25401762 17994087 20798317 19859648 24508170 15632085 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 1969491 12537569 21719571 20966253 12364791 14747013 17210077 24438588 21835214 24945155
EMBL
BABH01032611
NWSH01003020
PCG67105.1
KZ150069
PZC74080.1
AK401300
+ More
BAM17922.1 KP657638 RSAL01000006 AKL78863.1 RVE54315.1 GAIX01004181 JAA88379.1 BABH01007020 KQ461108 KPJ09262.1 AGBW02012593 OWR44772.1 NEVH01002994 PNF40921.1 KQ971327 EEZ99821.1 KK852685 KDR18484.1 CCAG010007360 EZ423398 ADD19674.1 GALX01003841 JAB64625.1 KQ435762 KOX75490.1 CP012528 ALC49436.1 JXJN01009401 HG965789 PYGN01000753 CDO39385.1 PSN41298.1 GECU01023633 JAS84073.1 BT126649 AEE61612.1 CVRI01000001 CRK86464.1 GEZM01081263 GEZM01081262 JAV61728.1 KU932410 APA34046.1 GBXI01000554 JAD13738.1 GAMC01006052 JAC00504.1 GBXI01011834 JAD02458.1 GDHF01015334 JAI36980.1 GAKP01012982 JAC45970.1 GDHF01009632 JAI42682.1 GAKP01012981 JAC45971.1 JRES01000777 KNC28415.1 GECU01020177 JAS87529.1 KY681699 AVD29897.1 GAHY01000111 JAA77399.1 GDKW01001673 JAI54922.1 KC445137 AGJ70738.1 KQ414663 KOC65354.1 GECZ01026871 JAS42898.1 KQ976491 KYM83318.1 CH478033 EAT34276.1 GECZ01018741 JAS51028.1 AY623406 AAV31915.1 EAT34277.1 KF021980 AGR65729.1 KQ978379 KYM94490.1 GBHO01033421 GBHO01005375 GBHO01005374 GBHO01005369 GBRD01014272 GBRD01011329 JAG10183.1 JAG38229.1 JAG38230.1 JAG38235.1 JAG51554.1 KQ982068 KYQ60545.1 CH963925 EDW78328.1 GFDL01000119 JAV34926.1 CH933810 EDW07386.1 CH940660 EDW58048.1 OUUW01000011 SPP86775.1 JR039080 AEY58519.1 KRF78088.1 KZ288311 PBC28530.1 CH916370 EDV99532.1 KRF94024.1 KQ459594 KPI96122.1 GL450772 EFN80494.1 GFDL01000209 JAV34836.1 KQ981622 KYN39215.1 FJ821026 ACN94650.1 KK107374 EZA51576.1 CH379064 EAL31931.1 CH479200 EDW29761.1 CM000162 EDX01973.1 AE014298 AHN59592.1 X52759 AY060701 BT099804 ACV91641.1 AHN59591.1 GFDF01001163 JAV12921.1 GEDC01022402 JAS14896.1 GL888197 EGI65384.1 UFQS01000234 UFQT01000234 SSX01768.1 SSX22146.1 CH954180 EDV46050.1 BT023772 AAZ41780.1 KRK06487.1 CH480851 EDW51385.1 KC344400 AGY46258.1 SPP86774.1 AAAB01008859 EAA08219.5 ATLV01013858 KE524905 KFB38209.1 HQ851380 AEV89758.1 GECL01003697 JAP02427.1 GL442742 EFN63125.1 EGK96917.1 APCN01000435 GAPW01002121 GAPW01002120 GAPW01002119 JAC11478.1 KQS29805.1
BAM17922.1 KP657638 RSAL01000006 AKL78863.1 RVE54315.1 GAIX01004181 JAA88379.1 BABH01007020 KQ461108 KPJ09262.1 AGBW02012593 OWR44772.1 NEVH01002994 PNF40921.1 KQ971327 EEZ99821.1 KK852685 KDR18484.1 CCAG010007360 EZ423398 ADD19674.1 GALX01003841 JAB64625.1 KQ435762 KOX75490.1 CP012528 ALC49436.1 JXJN01009401 HG965789 PYGN01000753 CDO39385.1 PSN41298.1 GECU01023633 JAS84073.1 BT126649 AEE61612.1 CVRI01000001 CRK86464.1 GEZM01081263 GEZM01081262 JAV61728.1 KU932410 APA34046.1 GBXI01000554 JAD13738.1 GAMC01006052 JAC00504.1 GBXI01011834 JAD02458.1 GDHF01015334 JAI36980.1 GAKP01012982 JAC45970.1 GDHF01009632 JAI42682.1 GAKP01012981 JAC45971.1 JRES01000777 KNC28415.1 GECU01020177 JAS87529.1 KY681699 AVD29897.1 GAHY01000111 JAA77399.1 GDKW01001673 JAI54922.1 KC445137 AGJ70738.1 KQ414663 KOC65354.1 GECZ01026871 JAS42898.1 KQ976491 KYM83318.1 CH478033 EAT34276.1 GECZ01018741 JAS51028.1 AY623406 AAV31915.1 EAT34277.1 KF021980 AGR65729.1 KQ978379 KYM94490.1 GBHO01033421 GBHO01005375 GBHO01005374 GBHO01005369 GBRD01014272 GBRD01011329 JAG10183.1 JAG38229.1 JAG38230.1 JAG38235.1 JAG51554.1 KQ982068 KYQ60545.1 CH963925 EDW78328.1 GFDL01000119 JAV34926.1 CH933810 EDW07386.1 CH940660 EDW58048.1 OUUW01000011 SPP86775.1 JR039080 AEY58519.1 KRF78088.1 KZ288311 PBC28530.1 CH916370 EDV99532.1 KRF94024.1 KQ459594 KPI96122.1 GL450772 EFN80494.1 GFDL01000209 JAV34836.1 KQ981622 KYN39215.1 FJ821026 ACN94650.1 KK107374 EZA51576.1 CH379064 EAL31931.1 CH479200 EDW29761.1 CM000162 EDX01973.1 AE014298 AHN59592.1 X52759 AY060701 BT099804 ACV91641.1 AHN59591.1 GFDF01001163 JAV12921.1 GEDC01022402 JAS14896.1 GL888197 EGI65384.1 UFQS01000234 UFQT01000234 SSX01768.1 SSX22146.1 CH954180 EDV46050.1 BT023772 AAZ41780.1 KRK06487.1 CH480851 EDW51385.1 KC344400 AGY46258.1 SPP86774.1 AAAB01008859 EAA08219.5 ATLV01013858 KE524905 KFB38209.1 HQ851380 AEV89758.1 GECL01003697 JAP02427.1 GL442742 EFN63125.1 EGK96917.1 APCN01000435 GAPW01002121 GAPW01002120 GAPW01002119 JAC11478.1 KQS29805.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000053240
UP000007151
UP000235965
+ More
UP000007266 UP000027135 UP000092444 UP000078200 UP000053105 UP000092553 UP000092460 UP000092445 UP000245037 UP000092443 UP000183832 UP000037069 UP000095301 UP000053825 UP000192221 UP000078540 UP000008820 UP000078542 UP000075809 UP000007798 UP000009192 UP000008792 UP000268350 UP000242457 UP000001070 UP000005203 UP000053268 UP000008237 UP000078541 UP000053097 UP000001819 UP000008744 UP000002282 UP000000803 UP000007755 UP000008711 UP000001292 UP000075902 UP000076407 UP000075882 UP000007062 UP000030765 UP000000311 UP000075903 UP000075840
UP000007266 UP000027135 UP000092444 UP000078200 UP000053105 UP000092553 UP000092460 UP000092445 UP000245037 UP000092443 UP000183832 UP000037069 UP000095301 UP000053825 UP000192221 UP000078540 UP000008820 UP000078542 UP000075809 UP000007798 UP000009192 UP000008792 UP000268350 UP000242457 UP000001070 UP000005203 UP000053268 UP000008237 UP000078541 UP000053097 UP000001819 UP000008744 UP000002282 UP000000803 UP000007755 UP000008711 UP000001292 UP000075902 UP000076407 UP000075882 UP000007062 UP000030765 UP000000311 UP000075903 UP000075840
Interpro
Gene 3D
ProteinModelPortal
H9J441
A0A2A4J4U6
A0A2W1BMY2
I4DJ32
A0A0G3VIA2
S4PCI7
+ More
H9ITV0 A0A194QUW7 A0A212ETE4 A0A2J7RJB5 D6WFH1 A0A067R5C5 D3TPP6 V5G3M6 A0A1A9UHR5 A0A0M9A1M4 A0A0M5J396 A0A1B0B749 A0A1B0AIW9 A0A068TK10 A0A1A9Y1D7 A0A1B6IB03 J3JTU0 A0A1J1HER3 A0A1Y1KMB3 A0A1I9WLM6 A0A0A1XT45 W8BUA3 A0A0A1WVL5 A0A0K8VEK6 A0A034VVT9 A0A0K8VUW5 A0A034VRN0 A0A0L0C841 A0A1B6IKT6 A0A2L1DH34 R4G5A5 A0A1I8NDX3 A0A0P4VPJ5 M9V250 A0A0L7R3F9 A0A1W4V0I0 A0A1B6EY34 A0A195BGP1 Q16J32 A0A1B6FLE3 Q2KQ97 S5NFE0 A0A1W4UNL4 A0A195C1J9 A0A0A9WUK9 A0A151XJR8 B4N1T9 A0A1Q3G582 B4L4A8 B4MCX6 A0A3B0JXI0 V9IDX6 A0A0Q9VZ95 A0A2A3E9Y7 B4JIW8 A0A088A2A8 A0A0Q9WMW7 A0A194PS35 E2BUV7 A0A1Q3G4Z2 A0A151JWT9 D0QWE3 A0A026W7I7 Q29H28 B4H0G6 B4PYB1 X2JJG8 P20478 C8VV58 X2JDA5 A0A1L8E2I2 A0A1B6CN61 F4WK81 A0A336LVS2 B3NUD5 P20478-3 Q494K4 A0A0R1EIC3 B4IK13 A0A0G2QRP6 A0A3B0JXJ2 A0A182U6F2 A0A182XIC8 A0A182LFG0 Q7QBX9 A0A084VJR5 G9C5D9 A0A0V0G2W8 E2ATW3 A0A182V4E1 F5HK76 A0A182HTB3 A0A023EQU3 A0A0Q5T3A2
H9ITV0 A0A194QUW7 A0A212ETE4 A0A2J7RJB5 D6WFH1 A0A067R5C5 D3TPP6 V5G3M6 A0A1A9UHR5 A0A0M9A1M4 A0A0M5J396 A0A1B0B749 A0A1B0AIW9 A0A068TK10 A0A1A9Y1D7 A0A1B6IB03 J3JTU0 A0A1J1HER3 A0A1Y1KMB3 A0A1I9WLM6 A0A0A1XT45 W8BUA3 A0A0A1WVL5 A0A0K8VEK6 A0A034VVT9 A0A0K8VUW5 A0A034VRN0 A0A0L0C841 A0A1B6IKT6 A0A2L1DH34 R4G5A5 A0A1I8NDX3 A0A0P4VPJ5 M9V250 A0A0L7R3F9 A0A1W4V0I0 A0A1B6EY34 A0A195BGP1 Q16J32 A0A1B6FLE3 Q2KQ97 S5NFE0 A0A1W4UNL4 A0A195C1J9 A0A0A9WUK9 A0A151XJR8 B4N1T9 A0A1Q3G582 B4L4A8 B4MCX6 A0A3B0JXI0 V9IDX6 A0A0Q9VZ95 A0A2A3E9Y7 B4JIW8 A0A088A2A8 A0A0Q9WMW7 A0A194PS35 E2BUV7 A0A1Q3G4Z2 A0A151JWT9 D0QWE3 A0A026W7I7 Q29H28 B4H0G6 B4PYB1 X2JJG8 P20478 C8VV58 X2JDA5 A0A1L8E2I2 A0A1B6CN61 F4WK81 A0A336LVS2 B3NUD5 P20478-3 Q494K4 A0A0R1EIC3 B4IK13 A0A0G2QRP6 A0A3B0JXJ2 A0A182U6F2 A0A182XIC8 A0A182LFG0 Q7QBX9 A0A084VJR5 G9C5D9 A0A0V0G2W8 E2ATW3 A0A182V4E1 F5HK76 A0A182HTB3 A0A023EQU3 A0A0Q5T3A2
PDB
2QC8
E-value=1.10236e-126,
Score=1160
Ontologies
PATHWAY
00220
Arginine biosynthesis - Bombyx mori (domestic silkworm)
00250 Alanine, aspartate and glutamate metabolism - Bombyx mori (domestic silkworm)
00630 Glyoxylate and dicarboxylate metabolism - Bombyx mori (domestic silkworm)
00910 Nitrogen metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
01230 Biosynthesis of amino acids - Bombyx mori (domestic silkworm)
00250 Alanine, aspartate and glutamate metabolism - Bombyx mori (domestic silkworm)
00630 Glyoxylate and dicarboxylate metabolism - Bombyx mori (domestic silkworm)
00910 Nitrogen metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
01230 Biosynthesis of amino acids - Bombyx mori (domestic silkworm)
GO
Topology
Subcellular location
Cytoplasm
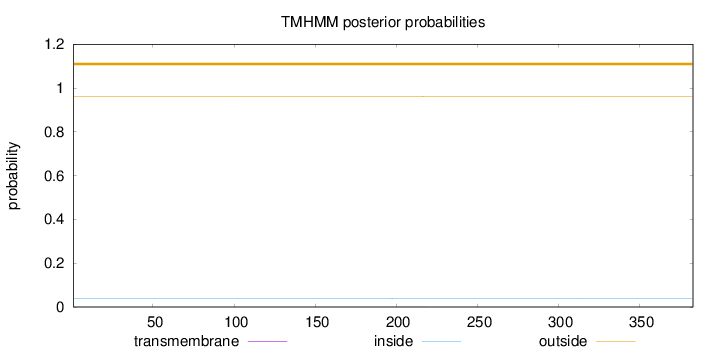
Length:
383
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00486
Exp number, first 60 AAs:
0.0002
Total prob of N-in:
0.03947
outside
1 - 383
Population Genetic Test Statistics
Pi
212.813192
Theta
156.671089
Tajima's D
1.132225
CLR
0.24629
CSRT
0.698065096745163
Interpretation
Uncertain
