Gene
KWMTBOMO12076 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA004331
Annotation
FK506-binding_protein_[Bombyx_mori]
Full name
Peptidylprolyl isomerase
Location in the cell
Cytoplasmic Reliability : 2.59
Sequence
CDS
ATGGGTGTTACTGTTGAAACTATCAGTCCAGGAGACGAGTCGACCTACCCCAAATCAGGACAGACTGTAGTTGTTCACTACACTGGAACACTTACGAACGGCAAAAAGTTTGACTCATCGCGTGACCGTGGCAAGCCCTTCAAGTTCAGGATCGGAAAATCTGAAGTAATCCGTGGCTGGGATGAGGGTGTGGCAAAGATGTCTGTAGGTGAGCGTGCCAAATTAACCTGCTCTCCTGACTATGCTTACGGTCAGCAAGGCCACCCTGGAGTCATACCTCCCAACTCTACACTTATTTTCGATGTAGAACTTCTACGTCTCGAATAA
Protein
MGVTVETISPGDESTYPKSGQTVVVHYTGTLTNGKKFDSSRDRGKPFKFRIGKSEVIRGWDEGVAKMSVGERAKLTCSPDYAYGQQGHPGVIPPNSTLIFDVELLRLE
Summary
Catalytic Activity
[protein]-peptidylproline (omega=180) = [protein]-peptidylproline (omega=0)
Feature
chain Peptidylprolyl isomerase
Uniprot
Q1HQ55
A0A2A4IRR3
A0A2H1VP43
A0FLQ3
N0ABL6
A0A2W1BHQ1
+ More
A0A0K8TCJ7 A0A212EME2 Q9U4Z3 A0A3S2P412 I7A0Q0 A0A194QRP8 I4DJR7 A0A0B5L9T6 S4PWZ3 Q966Y4 A0A0L7LTK4 Q6ZYL1 Q1HPH9 A0A0C9QYZ9 A0A131X8H2 D6WPQ3 A0A224YY99 A0A131YFN9 L7LWX7 E9HIY4 A0A0A9YSJ5 A0A0P5VEX5 V5GZ02 T1K9X4 A0A0N1PHX4 A0A0P4VI13 A0A0V0G525 R4FPA8 N0A6P9 W5L0I6 A0A1D2NIT1 A0A212FAP0 Q6PBJ8 A2I4A2 A0A194Q572 A0A171A5P3 A0A023F9G1 A0A069DVI6 S4PIK7 D1LX14 S4RXU0 A0A2A3EBC3 V9IL23 A0A088A2S8 A0A2R5LCK5 E3TDN4 E3TEM2 A0A1B6L1M3 A0A224XXJ4 A0A0L7KPB2 A0A226F2D1 A0A131Y1M2 A0A1V9XJD7 A0A087SZU3 A0A293M4E1 A0A1Y1K7T0 A0A1Z5LEU4 A0A158N9L0 A0A158N9L1 A0A147B8D9 A0A1B6CGG9 A0A1D1Z3H1 W5NIS8 C1BLM8 A0A1B6EME9 A0A3P8UWL0 A0A3Q3B3M7 A0A3P8YE67 A0A0P4XZB4 A0A3B4CGS4 A0A1B6EXS1 A0A1B6HRK1 A0A0K8REH0 K7J5Z5 Q8AVL7 U5EM76 A0A2A4JXM1 A0A2H1W8K8 A0A3Q2QQK2 A0A1W7RAH4 A0A3B1IZT9 Q642J1 Q5XG46 C1BM90 A0A0E9XJQ2 B5DGA2 A0A3Q3EJQ7 A0A1A7YNS7 A0A1A8FAL2 A0A0T6B6A5 A0A3B3TFB8 J3JUE3 A0A2I4C694 A0A2W1B956
A0A0K8TCJ7 A0A212EME2 Q9U4Z3 A0A3S2P412 I7A0Q0 A0A194QRP8 I4DJR7 A0A0B5L9T6 S4PWZ3 Q966Y4 A0A0L7LTK4 Q6ZYL1 Q1HPH9 A0A0C9QYZ9 A0A131X8H2 D6WPQ3 A0A224YY99 A0A131YFN9 L7LWX7 E9HIY4 A0A0A9YSJ5 A0A0P5VEX5 V5GZ02 T1K9X4 A0A0N1PHX4 A0A0P4VI13 A0A0V0G525 R4FPA8 N0A6P9 W5L0I6 A0A1D2NIT1 A0A212FAP0 Q6PBJ8 A2I4A2 A0A194Q572 A0A171A5P3 A0A023F9G1 A0A069DVI6 S4PIK7 D1LX14 S4RXU0 A0A2A3EBC3 V9IL23 A0A088A2S8 A0A2R5LCK5 E3TDN4 E3TEM2 A0A1B6L1M3 A0A224XXJ4 A0A0L7KPB2 A0A226F2D1 A0A131Y1M2 A0A1V9XJD7 A0A087SZU3 A0A293M4E1 A0A1Y1K7T0 A0A1Z5LEU4 A0A158N9L0 A0A158N9L1 A0A147B8D9 A0A1B6CGG9 A0A1D1Z3H1 W5NIS8 C1BLM8 A0A1B6EME9 A0A3P8UWL0 A0A3Q3B3M7 A0A3P8YE67 A0A0P4XZB4 A0A3B4CGS4 A0A1B6EXS1 A0A1B6HRK1 A0A0K8REH0 K7J5Z5 Q8AVL7 U5EM76 A0A2A4JXM1 A0A2H1W8K8 A0A3Q2QQK2 A0A1W7RAH4 A0A3B1IZT9 Q642J1 Q5XG46 C1BM90 A0A0E9XJQ2 B5DGA2 A0A3Q3EJQ7 A0A1A7YNS7 A0A1A8FAL2 A0A0T6B6A5 A0A3B3TFB8 J3JUE3 A0A2I4C694 A0A2W1B956
EC Number
5.2.1.8
Pubmed
19121390
23617895
28756777
22118469
10620045
26354079
+ More
22651552 23622113 26227816 26131772 28049606 18362917 19820115 28797301 26830274 25576852 21292972 25401762 26823975 27129103 25329095 27289101 23594743 25474469 26334808 20634964 28327890 28004739 28528879 21347285 24487278 25069045 20075255 25613341 19878547 29240929 22516182
22651552 23622113 26227816 26131772 28049606 18362917 19820115 28797301 26830274 25576852 21292972 25401762 26823975 27129103 25329095 27289101 23594743 25474469 26334808 20634964 28327890 28004739 28528879 21347285 24487278 25069045 20075255 25613341 19878547 29240929 22516182
EMBL
BABH01032597
DQ443197
ABF51286.1
NWSH01008741
PCG62431.1
ODYU01003437
+ More
SOQ42212.1 EF077224 ABK15648.1 KC481239 AGK45315.1 KZ150194 PZC72356.1 GBRD01002582 JAG63239.1 AGBW02013852 OWR42653.1 AF117595 AAF16717.1 RSAL01000029 RVE51807.1 JN546016 AFN85815.1 KQ461181 KPJ07655.1 AK401535 KQ459460 BAM18157.1 KPJ00825.1 KM923785 AJG43844.1 GAIX01005938 JAA86622.1 AJ278329 CAC38784.1 JTDY01000115 KOB78772.1 AJ634777 CAG25527.1 DQ443423 ABF51512.1 GBZX01002775 JAG89965.1 GEFH01004797 JAP63784.1 KQ971354 EFA06853.1 GFPF01007808 MAA18954.1 GEDV01011267 JAP77290.1 GACK01008719 JAA56315.1 GL732658 EFX68309.1 GBHO01011079 GBRD01002950 GDHC01011564 JAG32525.1 JAG62871.1 JAQ07065.1 GDIP01100803 GDIQ01037475 JAM02912.1 JAN57262.1 GALX01001349 JAB67117.1 CAEY01001893 KQ460009 KPJ18471.1 GDKW01002230 JAI54365.1 GECL01002975 JAP03149.1 GAHY01000974 JAA76536.1 KC481240 AGK45316.1 LJIJ01000029 ODN05147.1 AGBW02009448 OWR50789.1 CT737131 BC059682 AAH59682.1 EF070605 ABM55671.1 KPJ00692.1 GEMB01001474 GEMB01001473 JAS01683.1 GBBI01000576 JAC18136.1 GBGD01003565 JAC85324.1 GAIX01000169 JAA92391.1 GU075991 ACY92520.1 KZ288293 PBC28990.1 JR049571 AEY61004.1 GGLE01003128 MBY07254.1 GU588464 ADO28420.1 GU588804 ADO28758.1 GEBQ01022543 JAT17434.1 GFTR01001768 JAW14658.1 JTDY01008009 KOB64799.1 LNIX01000001 OXA63953.1 GEFM01003398 JAP72398.1 MNPL01009540 OQR73660.1 KK112716 KFM58382.1 GFWV01010330 MAA35059.1 GEZM01092425 JAV56561.1 GFJQ02001038 JAW05932.1 ADTU01009691 GEIB01001034 JAR87038.1 GEDC01024754 GEDC01009146 JAS12544.1 JAS28152.1 GDJX01006564 JAT61372.1 AHAT01027735 AHAT01027736 AHAT01027737 BT075507 ACO09931.1 GECZ01030680 JAS39089.1 GDIP01234754 JAI88647.1 GECZ01027015 JAS42754.1 GECU01030441 JAS77265.1 GADI01004934 JAA68874.1 BC041748 AAH41748.1 GANO01001136 JAB58735.1 NWSH01000415 PCG76549.1 ODYU01007025 SOQ49400.1 GFAH01000238 JAV48151.1 BX927368 BC081522 AAH81522.1 BC084619 BC086462 AAH84619.1 AAH86462.1 BT075719 ACO10143.1 GBXM01005698 JAI02880.1 BT043661 ACH70776.1 HADW01019090 HADX01009616 SBP31848.1 HAEB01009585 HAEC01014134 SBQ56112.1 LJIG01009607 KRT82741.1 BT126856 AEE61818.1 KZ150208 PZC72229.1
SOQ42212.1 EF077224 ABK15648.1 KC481239 AGK45315.1 KZ150194 PZC72356.1 GBRD01002582 JAG63239.1 AGBW02013852 OWR42653.1 AF117595 AAF16717.1 RSAL01000029 RVE51807.1 JN546016 AFN85815.1 KQ461181 KPJ07655.1 AK401535 KQ459460 BAM18157.1 KPJ00825.1 KM923785 AJG43844.1 GAIX01005938 JAA86622.1 AJ278329 CAC38784.1 JTDY01000115 KOB78772.1 AJ634777 CAG25527.1 DQ443423 ABF51512.1 GBZX01002775 JAG89965.1 GEFH01004797 JAP63784.1 KQ971354 EFA06853.1 GFPF01007808 MAA18954.1 GEDV01011267 JAP77290.1 GACK01008719 JAA56315.1 GL732658 EFX68309.1 GBHO01011079 GBRD01002950 GDHC01011564 JAG32525.1 JAG62871.1 JAQ07065.1 GDIP01100803 GDIQ01037475 JAM02912.1 JAN57262.1 GALX01001349 JAB67117.1 CAEY01001893 KQ460009 KPJ18471.1 GDKW01002230 JAI54365.1 GECL01002975 JAP03149.1 GAHY01000974 JAA76536.1 KC481240 AGK45316.1 LJIJ01000029 ODN05147.1 AGBW02009448 OWR50789.1 CT737131 BC059682 AAH59682.1 EF070605 ABM55671.1 KPJ00692.1 GEMB01001474 GEMB01001473 JAS01683.1 GBBI01000576 JAC18136.1 GBGD01003565 JAC85324.1 GAIX01000169 JAA92391.1 GU075991 ACY92520.1 KZ288293 PBC28990.1 JR049571 AEY61004.1 GGLE01003128 MBY07254.1 GU588464 ADO28420.1 GU588804 ADO28758.1 GEBQ01022543 JAT17434.1 GFTR01001768 JAW14658.1 JTDY01008009 KOB64799.1 LNIX01000001 OXA63953.1 GEFM01003398 JAP72398.1 MNPL01009540 OQR73660.1 KK112716 KFM58382.1 GFWV01010330 MAA35059.1 GEZM01092425 JAV56561.1 GFJQ02001038 JAW05932.1 ADTU01009691 GEIB01001034 JAR87038.1 GEDC01024754 GEDC01009146 JAS12544.1 JAS28152.1 GDJX01006564 JAT61372.1 AHAT01027735 AHAT01027736 AHAT01027737 BT075507 ACO09931.1 GECZ01030680 JAS39089.1 GDIP01234754 JAI88647.1 GECZ01027015 JAS42754.1 GECU01030441 JAS77265.1 GADI01004934 JAA68874.1 BC041748 AAH41748.1 GANO01001136 JAB58735.1 NWSH01000415 PCG76549.1 ODYU01007025 SOQ49400.1 GFAH01000238 JAV48151.1 BX927368 BC081522 AAH81522.1 BC084619 BC086462 AAH84619.1 AAH86462.1 BT075719 ACO10143.1 GBXM01005698 JAI02880.1 BT043661 ACH70776.1 HADW01019090 HADX01009616 SBP31848.1 HAEB01009585 HAEC01014134 SBQ56112.1 LJIG01009607 KRT82741.1 BT126856 AEE61818.1 KZ150208 PZC72229.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000283053
UP000053240
UP000053268
+ More
UP000037510 UP000007266 UP000000305 UP000015104 UP000018467 UP000094527 UP000000437 UP000245300 UP000242457 UP000005203 UP000221080 UP000198287 UP000192247 UP000054359 UP000005205 UP000018468 UP000265120 UP000264800 UP000265140 UP000261440 UP000002358 UP000265000 UP000087266 UP000261540 UP000192220
UP000037510 UP000007266 UP000000305 UP000015104 UP000018467 UP000094527 UP000000437 UP000245300 UP000242457 UP000005203 UP000221080 UP000198287 UP000192247 UP000054359 UP000005205 UP000018468 UP000265120 UP000264800 UP000265140 UP000261440 UP000002358 UP000265000 UP000087266 UP000261540 UP000192220
Pfam
PF00254 FKBP_C
ProteinModelPortal
Q1HQ55
A0A2A4IRR3
A0A2H1VP43
A0FLQ3
N0ABL6
A0A2W1BHQ1
+ More
A0A0K8TCJ7 A0A212EME2 Q9U4Z3 A0A3S2P412 I7A0Q0 A0A194QRP8 I4DJR7 A0A0B5L9T6 S4PWZ3 Q966Y4 A0A0L7LTK4 Q6ZYL1 Q1HPH9 A0A0C9QYZ9 A0A131X8H2 D6WPQ3 A0A224YY99 A0A131YFN9 L7LWX7 E9HIY4 A0A0A9YSJ5 A0A0P5VEX5 V5GZ02 T1K9X4 A0A0N1PHX4 A0A0P4VI13 A0A0V0G525 R4FPA8 N0A6P9 W5L0I6 A0A1D2NIT1 A0A212FAP0 Q6PBJ8 A2I4A2 A0A194Q572 A0A171A5P3 A0A023F9G1 A0A069DVI6 S4PIK7 D1LX14 S4RXU0 A0A2A3EBC3 V9IL23 A0A088A2S8 A0A2R5LCK5 E3TDN4 E3TEM2 A0A1B6L1M3 A0A224XXJ4 A0A0L7KPB2 A0A226F2D1 A0A131Y1M2 A0A1V9XJD7 A0A087SZU3 A0A293M4E1 A0A1Y1K7T0 A0A1Z5LEU4 A0A158N9L0 A0A158N9L1 A0A147B8D9 A0A1B6CGG9 A0A1D1Z3H1 W5NIS8 C1BLM8 A0A1B6EME9 A0A3P8UWL0 A0A3Q3B3M7 A0A3P8YE67 A0A0P4XZB4 A0A3B4CGS4 A0A1B6EXS1 A0A1B6HRK1 A0A0K8REH0 K7J5Z5 Q8AVL7 U5EM76 A0A2A4JXM1 A0A2H1W8K8 A0A3Q2QQK2 A0A1W7RAH4 A0A3B1IZT9 Q642J1 Q5XG46 C1BM90 A0A0E9XJQ2 B5DGA2 A0A3Q3EJQ7 A0A1A7YNS7 A0A1A8FAL2 A0A0T6B6A5 A0A3B3TFB8 J3JUE3 A0A2I4C694 A0A2W1B956
A0A0K8TCJ7 A0A212EME2 Q9U4Z3 A0A3S2P412 I7A0Q0 A0A194QRP8 I4DJR7 A0A0B5L9T6 S4PWZ3 Q966Y4 A0A0L7LTK4 Q6ZYL1 Q1HPH9 A0A0C9QYZ9 A0A131X8H2 D6WPQ3 A0A224YY99 A0A131YFN9 L7LWX7 E9HIY4 A0A0A9YSJ5 A0A0P5VEX5 V5GZ02 T1K9X4 A0A0N1PHX4 A0A0P4VI13 A0A0V0G525 R4FPA8 N0A6P9 W5L0I6 A0A1D2NIT1 A0A212FAP0 Q6PBJ8 A2I4A2 A0A194Q572 A0A171A5P3 A0A023F9G1 A0A069DVI6 S4PIK7 D1LX14 S4RXU0 A0A2A3EBC3 V9IL23 A0A088A2S8 A0A2R5LCK5 E3TDN4 E3TEM2 A0A1B6L1M3 A0A224XXJ4 A0A0L7KPB2 A0A226F2D1 A0A131Y1M2 A0A1V9XJD7 A0A087SZU3 A0A293M4E1 A0A1Y1K7T0 A0A1Z5LEU4 A0A158N9L0 A0A158N9L1 A0A147B8D9 A0A1B6CGG9 A0A1D1Z3H1 W5NIS8 C1BLM8 A0A1B6EME9 A0A3P8UWL0 A0A3Q3B3M7 A0A3P8YE67 A0A0P4XZB4 A0A3B4CGS4 A0A1B6EXS1 A0A1B6HRK1 A0A0K8REH0 K7J5Z5 Q8AVL7 U5EM76 A0A2A4JXM1 A0A2H1W8K8 A0A3Q2QQK2 A0A1W7RAH4 A0A3B1IZT9 Q642J1 Q5XG46 C1BM90 A0A0E9XJQ2 B5DGA2 A0A3Q3EJQ7 A0A1A7YNS7 A0A1A8FAL2 A0A0T6B6A5 A0A3B3TFB8 J3JUE3 A0A2I4C694 A0A2W1B956
PDB
5TB4
E-value=2.15885e-44,
Score=444
Ontologies
GO
PANTHER
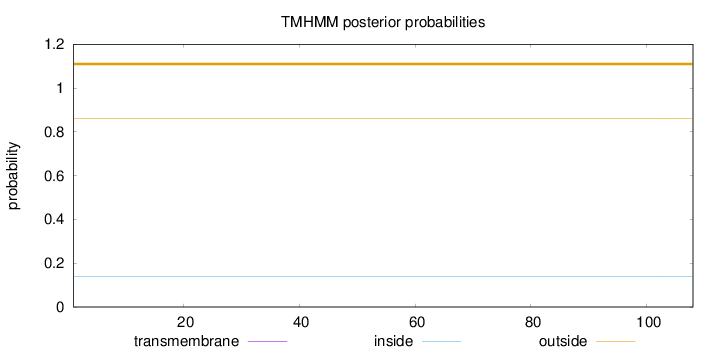
Topology
Length:
108
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00184
Exp number, first 60 AAs:
0.00148
Total prob of N-in:
0.13836
outside
1 - 108
Population Genetic Test Statistics
Pi
146.689239
Theta
139.808674
Tajima's D
-1.150457
CLR
0.348408
CSRT
0.112844357782111
Interpretation
Uncertain
