Gene
KWMTBOMO12060
Pre Gene Modal
BGIBMGA004301
Annotation
PREDICTED:_E3_ubiquitin-protein_ligase_RNF123-like_[Bombyx_mori]
Full name
E3 ubiquitin-protein ligase RNF123
Alternative Name
Kip1 ubiquitination-promoting complex protein 1
RING finger protein 123
RING-type E3 ubiquitin transferase RNF123
RING finger protein 123
RING-type E3 ubiquitin transferase RNF123
Location in the cell
Mitochondrial Reliability : 1.233
Sequence
CDS
ATGGCCGTGGTGTCTCGAGTGTGTATTCGTGGAGGCGGGGGGGCCAGCTTCGCGCGTCTTATAGCGCGCGGTCTCCCTGACACCGACAGGGTCCATTATTACGTAGCCTTGGCTCCCGTTGCGGGATGTCTCTTAGGGCTACTAGAGCCGAAATTACCACCGGAGCACTTAGATAAAGTGACGAAAGCGCTGATAATGGAACCGTTGTTCCGCATTGACGGTCTAGACTTCATGCTGGGAAAAACCAAGGACAAGAGCTTCTCCTTTTACCGATATCCGGAAGACGTGGCAGCGTCAGACTTGAAAGCGTTGGAGGTAGCGGTAGACCGTCTGCGAGTGGCACGCGATATAGTAACGAGGGCGCGGTCCCCGGGCTCCAGTGGGGCGCAGTCCGACTTGCTCTGCACCATATGCTACGCGAGACCCGCCGACACTGCCTTCGTACCGTGCGGACATCATTCTTGCAGAGCCTGCGTAATGCAGCATCTGCTCAACAGCAAACAATGTTTCTTCTGCAAAGCGGAAATCGATACAGTCCAAGAAATCGAGCCGGCTAACAATTAA
Protein
MAVVSRVCIRGGGGASFARLIARGLPDTDRVHYYVALAPVAGCLLGLLEPKLPPEHLDKVTKALIMEPLFRIDGLDFMLGKTKDKSFSFYRYPEDVAASDLKALEVAVDRLRVARDIVTRARSPGSSGAQSDLLCTICYARPADTAFVPCGHHSCRACVMQHLLNSKQCFFCKAEIDTVQEIEPANN
Summary
Description
Catalytic subunit of the KPC complex that acts as E3 ubiquitin-protein ligase. Required for poly-ubiquitination and proteasome-mediated degradation of CDKN1B during G1 phase of the cell cycle.
Catalytic Activity
S-ubiquitinyl-[E2 ubiquitin-conjugating enzyme]-L-cysteine + [acceptor protein]-L-lysine = [E2 ubiquitin-conjugating enzyme]-L-cysteine + N(6)-ubiquitinyl-[acceptor protein]-L-lysine.
Subunit
Component of the KPC complex composed of RNF123/KPC1 and UBAC1/KPC2. Interacts with UBAC1 and CDKN1B via its N-terminal domain.
Keywords
Acetylation
Alternative splicing
Complete proteome
Cytoplasm
Metal-binding
Methylation
Phosphoprotein
Reference proteome
Transferase
Ubl conjugation
Ubl conjugation pathway
Zinc
Zinc-finger
3D-structure
Polymorphism
Feature
chain E3 ubiquitin-protein ligase RNF123
splice variant In isoform 2.
sequence variant In dbSNP:rs2960546.
splice variant In isoform 2.
sequence variant In dbSNP:rs2960546.
Uniprot
A0A2W1B8F0
A0A2A4JZP7
A0A2H1W864
A0A0L7LBE9
A0A194QQ69
A0A194Q638
+ More
A0A232ETI7 K7IRA5 A0A2L2YC53 A0A2L2YC59 A0A088A2B9 A0A2A3E9Z9 E2AXN8 A0A1D2N598 A0A067RFY2 T1DCJ6 A0A1S3KH47 E2BUD9 A0A0B6XY99 A0A087U4A8 A0A158NPK7 A0A3L8D5W7 A0A026VY39 A0A0L7QZT7 F6TXP6 A0A195ELB7 G3TAQ7 G3UDJ9 F6TZ28 A0A1A6H0F7 A0A2T7NUJ8 A0A2K6BNB1 A0A2K5M3B5 A0A2K5JPN0 A0A2J8TGM6 A0A2K5ZHR4 G3S6F3 A0A2R9AZZ6 A0A1Z5LCS5 A0A2K6L3K7 T2MCA6 A0A2K6NBE4 H0X5J3 A0A1L8GP90 K7DHN4 K7AI08 V4A671 A0A0N0BH43 G1PTN3 A0A2J8TGR0 A0A226DAC8 A0A2K5EDK2 E0VGB7 A0A2K5S5G3 Q5XPI3-2 A0A2K5S5K0 A0A2K6BND4 A0A2K5ZHP9 A0A2I3LM12 A0A2K5M3D3 A0A2K5JPQ0 A0A2R9AW47 G3R4Y6 A0A1U7TU49 A0A2K6NBC6 A0A2K6L3M1 A0A2I3TW30 G1R5C7 U3DTN3 Q5XPI4 V8PA58 B3KU09 G3H8E8 A0A093PE73 A0A250YM69 A0A2K5UN43 G7ML89 G8F4N4 I0FU39 A0A2K5EDN6 D3ZXK7 A0A2K5S5K7 F1SPR6 C9JI97 E1C5W8 A0A3Q0CY04 I3LFZ7 A0A2R5LIJ3 A0A218UQG7 T1IK83
A0A232ETI7 K7IRA5 A0A2L2YC53 A0A2L2YC59 A0A088A2B9 A0A2A3E9Z9 E2AXN8 A0A1D2N598 A0A067RFY2 T1DCJ6 A0A1S3KH47 E2BUD9 A0A0B6XY99 A0A087U4A8 A0A158NPK7 A0A3L8D5W7 A0A026VY39 A0A0L7QZT7 F6TXP6 A0A195ELB7 G3TAQ7 G3UDJ9 F6TZ28 A0A1A6H0F7 A0A2T7NUJ8 A0A2K6BNB1 A0A2K5M3B5 A0A2K5JPN0 A0A2J8TGM6 A0A2K5ZHR4 G3S6F3 A0A2R9AZZ6 A0A1Z5LCS5 A0A2K6L3K7 T2MCA6 A0A2K6NBE4 H0X5J3 A0A1L8GP90 K7DHN4 K7AI08 V4A671 A0A0N0BH43 G1PTN3 A0A2J8TGR0 A0A226DAC8 A0A2K5EDK2 E0VGB7 A0A2K5S5G3 Q5XPI3-2 A0A2K5S5K0 A0A2K6BND4 A0A2K5ZHP9 A0A2I3LM12 A0A2K5M3D3 A0A2K5JPQ0 A0A2R9AW47 G3R4Y6 A0A1U7TU49 A0A2K6NBC6 A0A2K6L3M1 A0A2I3TW30 G1R5C7 U3DTN3 Q5XPI4 V8PA58 B3KU09 G3H8E8 A0A093PE73 A0A250YM69 A0A2K5UN43 G7ML89 G8F4N4 I0FU39 A0A2K5EDN6 D3ZXK7 A0A2K5S5K7 F1SPR6 C9JI97 E1C5W8 A0A3Q0CY04 I3LFZ7 A0A2R5LIJ3 A0A218UQG7 T1IK83
EC Number
2.3.2.27
Pubmed
28756777
26227816
26354079
28648823
20075255
26561354
+ More
20798317 27289101 24845553 26383154 21347285 30249741 24508170 19892987 22398555 22722832 28528879 24065732 25362486 27762356 16136131 23254933 21993624 20566863 15531880 15489334 21183079 24129315 25243066 16641997 15498874 14702039 11230166 16227581 21269460 22814378 24297900 21804562 28087693 17431167 22002653 25319552 15057822 19015242 22673903 15592404 30723633
20798317 27289101 24845553 26383154 21347285 30249741 24508170 19892987 22398555 22722832 28528879 24065732 25362486 27762356 16136131 23254933 21993624 20566863 15531880 15489334 21183079 24129315 25243066 16641997 15498874 14702039 11230166 16227581 21269460 22814378 24297900 21804562 28087693 17431167 22002653 25319552 15057822 19015242 22673903 15592404 30723633
EMBL
KZ150458
PZC70805.1
NWSH01000349
PCG77158.1
ODYU01006952
SOQ49271.1
+ More
JTDY01001920 KOB72541.1 KQ461181 KPJ07658.1 KQ459460 KPJ00829.1 NNAY01002280 OXU21637.1 IAAA01018209 LAA05746.1 IAAA01018210 IAAA01018211 LAA05752.1 KZ288311 PBC28540.1 GL443628 EFN61784.1 LJIJ01000233 ODN00116.1 KK852486 KDR22786.1 GAKT01000022 JAA93040.1 GL450640 EFN80666.1 HACG01001973 CEK48838.1 KK118079 KFM72197.1 ADTU01022570 ADTU01022571 ADTU01022572 ADTU01022573 QOIP01000012 RLU15862.1 KK107599 EZA48585.1 KQ414673 KOC64134.1 KQ978730 KYN29013.1 LZPO01058615 OBS71320.1 PZQS01000009 PVD24832.1 NDHI03003498 PNJ32205.1 CABD030021308 AJFE02064353 AJFE02064354 GFJQ02001952 JAW05018.1 HAAD01003298 CDG69530.1 AAQR03164084 AAQR03164085 AAQR03164086 CM004472 OCT85640.1 GABE01005829 NBAG03000218 JAA38910.1 PNI80599.1 AACZ04000315 GABC01003989 GABF01004529 JAA07349.1 JAA17616.1 KB202620 ESO88781.1 KQ435762 KOX75498.1 AAPE02059061 AAPE02059062 AAPE02059063 PNJ32211.1 LNIX01000028 OXA41687.1 DS235135 EEB12423.1 AY744153 BC057082 AHZZ02020133 AHZZ02020134 PNI80601.1 ADFV01173171 ADFV01173172 GAMT01005086 GAMS01010006 GAMP01001055 JAB06775.1 JAB13130.1 JAB51700.1 AY744152 AC099668 BC041145 BC057392 BC088801 BC130632 AF370367 AK022627 AK026968 AL136729 AZIM01000463 ETE70883.1 AK096353 BAG53271.1 JH000209 EGW08931.1 KL668904 KFW74716.1 GFFW01000071 JAV44717.1 AQIA01039642 JSUE03023699 JU476736 JU476737 CM001254 AFH33540.1 EHH16359.1 JH330954 EHH62248.1 JV047894 AFI37965.1 AEMK02000086 AADN05000356 DQIR01100801 HDA56277.1 GGLE01005021 MBY09147.1 MUZQ01000178 OWK56015.1 JH430443
JTDY01001920 KOB72541.1 KQ461181 KPJ07658.1 KQ459460 KPJ00829.1 NNAY01002280 OXU21637.1 IAAA01018209 LAA05746.1 IAAA01018210 IAAA01018211 LAA05752.1 KZ288311 PBC28540.1 GL443628 EFN61784.1 LJIJ01000233 ODN00116.1 KK852486 KDR22786.1 GAKT01000022 JAA93040.1 GL450640 EFN80666.1 HACG01001973 CEK48838.1 KK118079 KFM72197.1 ADTU01022570 ADTU01022571 ADTU01022572 ADTU01022573 QOIP01000012 RLU15862.1 KK107599 EZA48585.1 KQ414673 KOC64134.1 KQ978730 KYN29013.1 LZPO01058615 OBS71320.1 PZQS01000009 PVD24832.1 NDHI03003498 PNJ32205.1 CABD030021308 AJFE02064353 AJFE02064354 GFJQ02001952 JAW05018.1 HAAD01003298 CDG69530.1 AAQR03164084 AAQR03164085 AAQR03164086 CM004472 OCT85640.1 GABE01005829 NBAG03000218 JAA38910.1 PNI80599.1 AACZ04000315 GABC01003989 GABF01004529 JAA07349.1 JAA17616.1 KB202620 ESO88781.1 KQ435762 KOX75498.1 AAPE02059061 AAPE02059062 AAPE02059063 PNJ32211.1 LNIX01000028 OXA41687.1 DS235135 EEB12423.1 AY744153 BC057082 AHZZ02020133 AHZZ02020134 PNI80601.1 ADFV01173171 ADFV01173172 GAMT01005086 GAMS01010006 GAMP01001055 JAB06775.1 JAB13130.1 JAB51700.1 AY744152 AC099668 BC041145 BC057392 BC088801 BC130632 AF370367 AK022627 AK026968 AL136729 AZIM01000463 ETE70883.1 AK096353 BAG53271.1 JH000209 EGW08931.1 KL668904 KFW74716.1 GFFW01000071 JAV44717.1 AQIA01039642 JSUE03023699 JU476736 JU476737 CM001254 AFH33540.1 EHH16359.1 JH330954 EHH62248.1 JV047894 AFI37965.1 AEMK02000086 AADN05000356 DQIR01100801 HDA56277.1 GGLE01005021 MBY09147.1 MUZQ01000178 OWK56015.1 JH430443
Proteomes
UP000218220
UP000037510
UP000053240
UP000053268
UP000215335
UP000002358
+ More
UP000005203 UP000242457 UP000000311 UP000094527 UP000027135 UP000085678 UP000008237 UP000054359 UP000005205 UP000279307 UP000053097 UP000053825 UP000002281 UP000078492 UP000007646 UP000092124 UP000245119 UP000233120 UP000233060 UP000233080 UP000233140 UP000001519 UP000240080 UP000233180 UP000233200 UP000005225 UP000186698 UP000002277 UP000030746 UP000053105 UP000001074 UP000198287 UP000233020 UP000009046 UP000233040 UP000000589 UP000028761 UP000189704 UP000001073 UP000008225 UP000005640 UP000001075 UP000053258 UP000233100 UP000006718 UP000009130 UP000002494 UP000008227 UP000000539 UP000189706 UP000197619
UP000005203 UP000242457 UP000000311 UP000094527 UP000027135 UP000085678 UP000008237 UP000054359 UP000005205 UP000279307 UP000053097 UP000053825 UP000002281 UP000078492 UP000007646 UP000092124 UP000245119 UP000233120 UP000233060 UP000233080 UP000233140 UP000001519 UP000240080 UP000233180 UP000233200 UP000005225 UP000186698 UP000002277 UP000030746 UP000053105 UP000001074 UP000198287 UP000233020 UP000009046 UP000233040 UP000000589 UP000028761 UP000189704 UP000001073 UP000008225 UP000005640 UP000001075 UP000053258 UP000233100 UP000006718 UP000009130 UP000002494 UP000008227 UP000000539 UP000189706 UP000197619
Pfam
PF00622 SPRY
Interpro
Gene 3D
ProteinModelPortal
A0A2W1B8F0
A0A2A4JZP7
A0A2H1W864
A0A0L7LBE9
A0A194QQ69
A0A194Q638
+ More
A0A232ETI7 K7IRA5 A0A2L2YC53 A0A2L2YC59 A0A088A2B9 A0A2A3E9Z9 E2AXN8 A0A1D2N598 A0A067RFY2 T1DCJ6 A0A1S3KH47 E2BUD9 A0A0B6XY99 A0A087U4A8 A0A158NPK7 A0A3L8D5W7 A0A026VY39 A0A0L7QZT7 F6TXP6 A0A195ELB7 G3TAQ7 G3UDJ9 F6TZ28 A0A1A6H0F7 A0A2T7NUJ8 A0A2K6BNB1 A0A2K5M3B5 A0A2K5JPN0 A0A2J8TGM6 A0A2K5ZHR4 G3S6F3 A0A2R9AZZ6 A0A1Z5LCS5 A0A2K6L3K7 T2MCA6 A0A2K6NBE4 H0X5J3 A0A1L8GP90 K7DHN4 K7AI08 V4A671 A0A0N0BH43 G1PTN3 A0A2J8TGR0 A0A226DAC8 A0A2K5EDK2 E0VGB7 A0A2K5S5G3 Q5XPI3-2 A0A2K5S5K0 A0A2K6BND4 A0A2K5ZHP9 A0A2I3LM12 A0A2K5M3D3 A0A2K5JPQ0 A0A2R9AW47 G3R4Y6 A0A1U7TU49 A0A2K6NBC6 A0A2K6L3M1 A0A2I3TW30 G1R5C7 U3DTN3 Q5XPI4 V8PA58 B3KU09 G3H8E8 A0A093PE73 A0A250YM69 A0A2K5UN43 G7ML89 G8F4N4 I0FU39 A0A2K5EDN6 D3ZXK7 A0A2K5S5K7 F1SPR6 C9JI97 E1C5W8 A0A3Q0CY04 I3LFZ7 A0A2R5LIJ3 A0A218UQG7 T1IK83
A0A232ETI7 K7IRA5 A0A2L2YC53 A0A2L2YC59 A0A088A2B9 A0A2A3E9Z9 E2AXN8 A0A1D2N598 A0A067RFY2 T1DCJ6 A0A1S3KH47 E2BUD9 A0A0B6XY99 A0A087U4A8 A0A158NPK7 A0A3L8D5W7 A0A026VY39 A0A0L7QZT7 F6TXP6 A0A195ELB7 G3TAQ7 G3UDJ9 F6TZ28 A0A1A6H0F7 A0A2T7NUJ8 A0A2K6BNB1 A0A2K5M3B5 A0A2K5JPN0 A0A2J8TGM6 A0A2K5ZHR4 G3S6F3 A0A2R9AZZ6 A0A1Z5LCS5 A0A2K6L3K7 T2MCA6 A0A2K6NBE4 H0X5J3 A0A1L8GP90 K7DHN4 K7AI08 V4A671 A0A0N0BH43 G1PTN3 A0A2J8TGR0 A0A226DAC8 A0A2K5EDK2 E0VGB7 A0A2K5S5G3 Q5XPI3-2 A0A2K5S5K0 A0A2K6BND4 A0A2K5ZHP9 A0A2I3LM12 A0A2K5M3D3 A0A2K5JPQ0 A0A2R9AW47 G3R4Y6 A0A1U7TU49 A0A2K6NBC6 A0A2K6L3M1 A0A2I3TW30 G1R5C7 U3DTN3 Q5XPI4 V8PA58 B3KU09 G3H8E8 A0A093PE73 A0A250YM69 A0A2K5UN43 G7ML89 G8F4N4 I0FU39 A0A2K5EDN6 D3ZXK7 A0A2K5S5K7 F1SPR6 C9JI97 E1C5W8 A0A3Q0CY04 I3LFZ7 A0A2R5LIJ3 A0A218UQG7 T1IK83
PDB
2MA6
E-value=4.50388e-13,
Score=176
Ontologies
GO
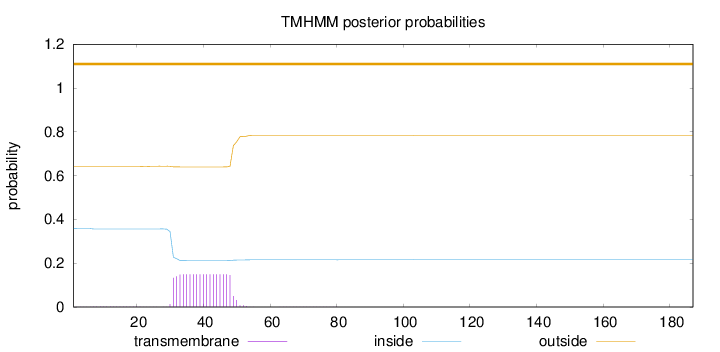
Topology
Subcellular location
Cytoplasm
Length:
187
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
2.80277000000001
Exp number, first 60 AAs:
2.79572
Total prob of N-in:
0.35808
outside
1 - 187
Population Genetic Test Statistics
Pi
79.429119
Theta
122.366672
Tajima's D
-0.717116
CLR
132.267746
CSRT
0.193040347982601
Interpretation
Uncertain
