Gene
KWMTBOMO12043
Pre Gene Modal
BGIBMGA004308
Annotation
PREDICTED:_uncharacterized_protein_LOC106135922_[Amyelois_transitella]
Location in the cell
PlasmaMembrane Reliability : 3.645
Sequence
CDS
ATGGGTAATGCCATTGGAAAATTTCTAACTGCGGGCAACGTTCAAGGTGCTGGCTCGGTTCTCGATAGAGTCATATCCCAGCCGAGCAGCGAAGACCACACCGTACTTTACAAATTAGCTGATTATAAGAAAGGCGGTCTACTTTTAGAGACCTACGCGAAAGGTGGCATGACAGCCGCTGAAAAGTTAATGAGAGACGAATTCGCTGCATACATGTACGGCGGAGGGAGGGGCCGGGTCATCAATCGTGCTGAATACTTGAGATGGAAGTTCAGAGACCAAGAACAGGTTGTACTTCCAATTGAAGCATCGCTCTCACCTTATGACCCGCTCGCCAAATGGGAAGATCACACCGCCTGTTGGCAGATGTGTTATAGAGGAGCTCTCGGCGAATCATTGCTTCATGTACTCATTATTTGTGATACTAAGATTCATACAAGATTAGCTCGAACTCTAGTAAAATGTTTTCCGAAGCTGTCACTAGATGTCGTTGAAGGTGAAGAATATTTAGGGGCAAGCTCTTTACATCTGGCAATAGCGTACAGCAATAACGAACTAGTACAAGATCTAGTTGAGGCCGGAGCTGATGTCAACCAGAGAGCCATTGGTAGCTTTTTCCTACCCCGCGACCAACAAAGAGTTCCGCCAGCGCGACAAACAAATTACGAAGGTCTGGCATACTTGGGCGAATACCCCCTAGCCTGGACGGCTTGTTGCGCGAACGAAGCAGTCTACAACCTCTTACTAGATTCCGGGGCAGATCCCGATGCTCAGGATTCGTTCGGGAACATGATACTACACATGGTGGTCGTTTGTGATAAACTGGACATGTTTGGATATGCCCTGCGTCACCCTAAAGTTCCGGCCAGTAACGGTCGAATGAATAAAGCAGGTTTCACCCCACTAACCTTGGCTTGTCAGTTGGGTCGGGCCTCTGTCTTCAGAGAGATGCTGGAGCTATCAGCCAGAGAGTTCTGGCGCTACTCCAACATTACGTGCTCCGCTTACCCTTTGAACGCTTTAGATACCTTGCTGCCTGATGGGCGAACAAATTGGAATTCGGCTTTATTCATTATACTGAACGGAACAAAGCAAGAACACTTGAACATGTTAGACGGCGGTATCATACAAAGGTTGCTTGAAGAAAAATGGAAGACTTTCGCCAGGACGAAATTCCTCAAACGTCTCCTCATACTGATGCTGCATCTTCTCCTGCTCTCCGTTTCTGTGTACCTACGTCACAGCAGCGCAGAAGCTGATGCCCACCCCAACTGGGGCTTGGAGATCAACGACGCTAGAAGCGGACTGAGACTTGCCAGTGAACTTGGGACTATATTGAGCACATTGTGTTATATTATACTGCAACAAGGCGACGAGCTAAAGAATCAAGGATTGGTCGCTTATTTTAAGCAGTTGATTCACGAGCCGGCCAAATTTATATTTTTGGCGTCAAACATCCTTGTGTTAGCGTGCATACCGGCCAGACTCCTTAAGGAGACCAACGTGGAGGAAGCCATACTGCTTTTCCTGCTGCCGGGATCGTGGTTTTTGTTGATGTTTTTTGCTGGCGCGGTGAAATTGACCGGTCCATTCGTGACAATGATCTACAGTATGATAACAGGGGACATGTTCACGTTCGGTATCATCTACTGTATTGTCCTCTTCGGCTTCTCGCAGTCATTCTACTTCTTGTATAGAGGTTTTCCAAACGTACAGTCGACTTTGTACTCGAGCTACCCCAGCACGTGGATGGCACTATTCCAAATCACCTTAGGAGATTACAGCTACTCGGATCTCAGTCAGACAACATACCCTAATCTCTCGAAGACTGTGTTCACGGTGTTCATGATATTCGTGCCAATTCTACTATTGAACATGTTGATCGCCATGATGGGGAACACGTACGCCCACGTCATTGAGCAATCCGAGAAGGAATGGGTCAAACAGTGGGCGAAAATTGTGGTATCACTAGAGCGTTCAGTGGCCCAGGATGACGCACATAAGTACTTGCAGGAGTATTCTATAGGACTTGGGCCTTCCGACGATCCGAGATACGAGCAACGAGCGGTCATGGTCATCAAGAGCAAAGCTAAAACTAGAGCGAAACAGAAGAAAGACGCTTTAACTAATTGGAAGCACGTCGGTAAAGTGACAATAGCAGAGTTGAGAAGACGCGGTATCAGCGGTGAAGAGTTACGTAGATTGATGTGGGGCCGTATTTCAATTTCGACACCAACAAAAGCTCCCTTGCCGCGAAGAGTCCCAGCCCCGCCTCCAGACTGCGTGGTTTCGTCAGACGTGGGTTTAGCTAGTGACGTAGGGAACGGTGTAGCACCAGCTCTCTCGTCAGCTCTCAACGTCATGGCCTTCACTCACGAATTAGACATTGGCACTACTGGTTCTGATCAAAAACAAACTACACCGGACTTGTTGGTCAACGGCAAAACTTCCAATGCCCCAATCTTAACTACCGGTACCCAAATGCCAAAAGTTTCAAGCAAAACTCTCGGTACACCAGTGGCACGTATTGAAGCATCTTCACTTAAGACGCCTTTGGATGTTAAATTAAACCCAACAACATTGACACAGTCCACGGTTCAGGGTAATATAGGTAATGTCAATATTCCTATAAGTGGAGTGAAAATAACAACAAATGTTGGACAAAACTCCGCGGTAGTACCTGAGATTCAACAGTCGAATGAAAATCAGAAACCTGAGCAAGTACACCAAGATTATCTGCGTGAACTCATAGTTCTTGCTGAGAAACCTGCAACCACTAATCTTGAACTCAAACAACTTGCTGAGAAAGCGGCTGATCTGAGAGATGTACCGGAAATTGATATTAACATCAATGTGAGTTAG
Protein
MGNAIGKFLTAGNVQGAGSVLDRVISQPSSEDHTVLYKLADYKKGGLLLETYAKGGMTAAEKLMRDEFAAYMYGGGRGRVINRAEYLRWKFRDQEQVVLPIEASLSPYDPLAKWEDHTACWQMCYRGALGESLLHVLIICDTKIHTRLARTLVKCFPKLSLDVVEGEEYLGASSLHLAIAYSNNELVQDLVEAGADVNQRAIGSFFLPRDQQRVPPARQTNYEGLAYLGEYPLAWTACCANEAVYNLLLDSGADPDAQDSFGNMILHMVVVCDKLDMFGYALRHPKVPASNGRMNKAGFTPLTLACQLGRASVFREMLELSAREFWRYSNITCSAYPLNALDTLLPDGRTNWNSALFIILNGTKQEHLNMLDGGIIQRLLEEKWKTFARTKFLKRLLILMLHLLLLSVSVYLRHSSAEADAHPNWGLEINDARSGLRLASELGTILSTLCYIILQQGDELKNQGLVAYFKQLIHEPAKFIFLASNILVLACIPARLLKETNVEEAILLFLLPGSWFLLMFFAGAVKLTGPFVTMIYSMITGDMFTFGIIYCIVLFGFSQSFYFLYRGFPNVQSTLYSSYPSTWMALFQITLGDYSYSDLSQTTYPNLSKTVFTVFMIFVPILLLNMLIAMMGNTYAHVIEQSEKEWVKQWAKIVVSLERSVAQDDAHKYLQEYSIGLGPSDDPRYEQRAVMVIKSKAKTRAKQKKDALTNWKHVGKVTIAELRRRGISGEELRRLMWGRISISTPTKAPLPRRVPAPPPDCVVSSDVGLASDVGNGVAPALSSALNVMAFTHELDIGTTGSDQKQTTPDLLVNGKTSNAPILTTGTQMPKVSSKTLGTPVARIEASSLKTPLDVKLNPTTLTQSTVQGNIGNVNIPISGVKITTNVGQNSAVVPEIQQSNENQKPEQVHQDYLRELIVLAEKPATTNLELKQLAEKAADLRDVPEIDININVS
Summary
Similarity
Belongs to the transient receptor (TC 1.A.4) family.
Uniprot
H9J469
A0A2W1BP56
A0A2A4JYR0
A0A212F2U8
A0A194Q759
A0A194QQN8
+ More
A0A2H1W9C4 F4WYD6 E2BHT3 A0A158NXN7 A0A151WPP2 A0A087ZPY6 D6X1X3 A0A195B0K6 A0A2D3C336 K7IWA0 A0A0L7RJR1 A0A195EVS4 E0W1R2 E2AXW0 A0A154NY96 A0A310SLY8 A0A232FJA6 T1HDJ1 A0A384THG1 A0A3L8E411 A0A195EDX7 A0A151INR2 U4TXE5 A0A1B0DB04 N6U8L1 A0A1I8MW96 A0A3B0KRT7 A0A026WJG3 A0A1A9UDZ9 A0A1B0BB69 Q29IL6 A0A0L0BL70 A0A1A9WL64 A0A1I8NP90 B3NXW2 B4I049 Q9W3W0 B4L526 B4PZ84 B4M701 A0A1W4UW54 B4JLU9 A0A1A9ZQC7 A0A0M4ERE6 A0A067RC69 A0A182YCS3 A0A182P182 A0A182RKY4 U5EG67 A0A182KAD1 A0A2D3C338 A0A1S4FM61 Q16WC0 A0A1Q3FSY0 B0X3V3 A0A182SDK7 A0A182GRZ1 A0A182N0L1 A0A182W5T4 A0A182LY80 A0A0A9Y583 A0A0M9A4X0 J9JUJ3 A0A336MUD6 A0A226DJ08 A0A1D2MQE6 A0A1J1HMG2 A0A0P6CKS2 E9FVS1 T1GKP7 A0A2P8Z3M5 A0A0K2T5U3 T1IMS0
A0A2H1W9C4 F4WYD6 E2BHT3 A0A158NXN7 A0A151WPP2 A0A087ZPY6 D6X1X3 A0A195B0K6 A0A2D3C336 K7IWA0 A0A0L7RJR1 A0A195EVS4 E0W1R2 E2AXW0 A0A154NY96 A0A310SLY8 A0A232FJA6 T1HDJ1 A0A384THG1 A0A3L8E411 A0A195EDX7 A0A151INR2 U4TXE5 A0A1B0DB04 N6U8L1 A0A1I8MW96 A0A3B0KRT7 A0A026WJG3 A0A1A9UDZ9 A0A1B0BB69 Q29IL6 A0A0L0BL70 A0A1A9WL64 A0A1I8NP90 B3NXW2 B4I049 Q9W3W0 B4L526 B4PZ84 B4M701 A0A1W4UW54 B4JLU9 A0A1A9ZQC7 A0A0M4ERE6 A0A067RC69 A0A182YCS3 A0A182P182 A0A182RKY4 U5EG67 A0A182KAD1 A0A2D3C338 A0A1S4FM61 Q16WC0 A0A1Q3FSY0 B0X3V3 A0A182SDK7 A0A182GRZ1 A0A182N0L1 A0A182W5T4 A0A182LY80 A0A0A9Y583 A0A0M9A4X0 J9JUJ3 A0A336MUD6 A0A226DJ08 A0A1D2MQE6 A0A1J1HMG2 A0A0P6CKS2 E9FVS1 T1GKP7 A0A2P8Z3M5 A0A0K2T5U3 T1IMS0
Pubmed
19121390
28756777
22118469
26354079
21719571
20798317
+ More
21347285 18362917 19820115 20075255 20566863 28648823 30249741 23537049 25315136 24508170 15632085 17994087 26108605 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17550304 24845553 25244985 17510324 26483478 25401762 27289101 21292972 29403074
21347285 18362917 19820115 20075255 20566863 28648823 30249741 23537049 25315136 24508170 15632085 17994087 26108605 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17550304 24845553 25244985 17510324 26483478 25401762 27289101 21292972 29403074
EMBL
BABH01032522
KZ150034
PZC74646.1
NWSH01000349
PCG77161.1
AGBW02010674
+ More
OWR48059.1 KQ459460 KPJ00835.1 KQ461181 KPJ07664.1 ODYU01007165 SOQ49705.1 GL888440 EGI60788.1 GL448324 EFN84766.1 ADTU01003256 ADTU01003257 ADTU01003258 ADTU01003259 KQ982851 KYQ49872.1 KQ971371 EFA10736.2 KQ976692 KYM77734.1 MF684873 ATU07274.1 KQ414579 KOC71074.1 KQ981958 KYN31994.1 AAZO01007084 DS235873 EEB19644.1 GL443736 EFN61724.1 KQ434782 KZC04593.1 KQ762788 OAD55522.1 NNAY01000165 OXU30387.1 ACPB03005165 KX249698 AOR81475.1 QOIP01000001 RLU27431.1 KQ979074 KYN23007.1 KQ976914 KYN07104.1 KB631698 ERL85467.1 AJVK01000574 AJVK01000575 AJVK01000576 AJVK01000577 APGK01038500 KB740960 ENN76991.1 OUUW01000011 SPP86638.1 KK107168 EZA56187.1 JXJN01011311 CH379063 EAL32637.1 JRES01001701 KNC20855.1 CH954180 EDV47413.1 CH480819 EDW52880.1 AE014298 AAF46203.1 CH933811 EDW06285.2 CM000162 EDX01051.1 CH940653 EDW62568.2 CH916371 EDV91710.1 CP012528 ALC48690.1 KK852801 KDR16309.1 GANO01003572 JAB56299.1 MF684872 ATU07273.1 CH477570 EAT38898.1 GFDL01004387 JAV30658.1 DS232323 EDS40030.1 JXUM01083500 KQ563380 KXJ73944.1 AXCM01006171 GBHO01017341 JAG26263.1 KQ435746 KOX76526.1 ABLF02035466 UFQT01002378 SSX33335.1 LNIX01000017 OXA45512.1 LJIJ01000688 ODM95297.1 CVRI01000011 CRK89237.1 GDIP01000670 LRGB01002849 JAN03046.1 KZS06144.1 GL732525 EFX88604.1 CAQQ02391715 PYGN01000212 PSN51083.1 HACA01004027 CDW21388.1 JH431094
OWR48059.1 KQ459460 KPJ00835.1 KQ461181 KPJ07664.1 ODYU01007165 SOQ49705.1 GL888440 EGI60788.1 GL448324 EFN84766.1 ADTU01003256 ADTU01003257 ADTU01003258 ADTU01003259 KQ982851 KYQ49872.1 KQ971371 EFA10736.2 KQ976692 KYM77734.1 MF684873 ATU07274.1 KQ414579 KOC71074.1 KQ981958 KYN31994.1 AAZO01007084 DS235873 EEB19644.1 GL443736 EFN61724.1 KQ434782 KZC04593.1 KQ762788 OAD55522.1 NNAY01000165 OXU30387.1 ACPB03005165 KX249698 AOR81475.1 QOIP01000001 RLU27431.1 KQ979074 KYN23007.1 KQ976914 KYN07104.1 KB631698 ERL85467.1 AJVK01000574 AJVK01000575 AJVK01000576 AJVK01000577 APGK01038500 KB740960 ENN76991.1 OUUW01000011 SPP86638.1 KK107168 EZA56187.1 JXJN01011311 CH379063 EAL32637.1 JRES01001701 KNC20855.1 CH954180 EDV47413.1 CH480819 EDW52880.1 AE014298 AAF46203.1 CH933811 EDW06285.2 CM000162 EDX01051.1 CH940653 EDW62568.2 CH916371 EDV91710.1 CP012528 ALC48690.1 KK852801 KDR16309.1 GANO01003572 JAB56299.1 MF684872 ATU07273.1 CH477570 EAT38898.1 GFDL01004387 JAV30658.1 DS232323 EDS40030.1 JXUM01083500 KQ563380 KXJ73944.1 AXCM01006171 GBHO01017341 JAG26263.1 KQ435746 KOX76526.1 ABLF02035466 UFQT01002378 SSX33335.1 LNIX01000017 OXA45512.1 LJIJ01000688 ODM95297.1 CVRI01000011 CRK89237.1 GDIP01000670 LRGB01002849 JAN03046.1 KZS06144.1 GL732525 EFX88604.1 CAQQ02391715 PYGN01000212 PSN51083.1 HACA01004027 CDW21388.1 JH431094
Proteomes
UP000005204
UP000218220
UP000007151
UP000053268
UP000053240
UP000007755
+ More
UP000008237 UP000005205 UP000075809 UP000005203 UP000007266 UP000078540 UP000002358 UP000053825 UP000078541 UP000009046 UP000000311 UP000076502 UP000215335 UP000015103 UP000279307 UP000078492 UP000078542 UP000030742 UP000092462 UP000019118 UP000095301 UP000268350 UP000053097 UP000078200 UP000092460 UP000001819 UP000037069 UP000091820 UP000095300 UP000008711 UP000001292 UP000000803 UP000009192 UP000002282 UP000008792 UP000192221 UP000001070 UP000092445 UP000092553 UP000027135 UP000076408 UP000075885 UP000075900 UP000075881 UP000008820 UP000002320 UP000075901 UP000069940 UP000249989 UP000075884 UP000075920 UP000075883 UP000053105 UP000007819 UP000198287 UP000094527 UP000183832 UP000076858 UP000000305 UP000015102 UP000245037
UP000008237 UP000005205 UP000075809 UP000005203 UP000007266 UP000078540 UP000002358 UP000053825 UP000078541 UP000009046 UP000000311 UP000076502 UP000215335 UP000015103 UP000279307 UP000078492 UP000078542 UP000030742 UP000092462 UP000019118 UP000095301 UP000268350 UP000053097 UP000078200 UP000092460 UP000001819 UP000037069 UP000091820 UP000095300 UP000008711 UP000001292 UP000000803 UP000009192 UP000002282 UP000008792 UP000192221 UP000001070 UP000092445 UP000092553 UP000027135 UP000076408 UP000075885 UP000075900 UP000075881 UP000008820 UP000002320 UP000075901 UP000069940 UP000249989 UP000075884 UP000075920 UP000075883 UP000053105 UP000007819 UP000198287 UP000094527 UP000183832 UP000076858 UP000000305 UP000015102 UP000245037
Interpro
Gene 3D
ProteinModelPortal
H9J469
A0A2W1BP56
A0A2A4JYR0
A0A212F2U8
A0A194Q759
A0A194QQN8
+ More
A0A2H1W9C4 F4WYD6 E2BHT3 A0A158NXN7 A0A151WPP2 A0A087ZPY6 D6X1X3 A0A195B0K6 A0A2D3C336 K7IWA0 A0A0L7RJR1 A0A195EVS4 E0W1R2 E2AXW0 A0A154NY96 A0A310SLY8 A0A232FJA6 T1HDJ1 A0A384THG1 A0A3L8E411 A0A195EDX7 A0A151INR2 U4TXE5 A0A1B0DB04 N6U8L1 A0A1I8MW96 A0A3B0KRT7 A0A026WJG3 A0A1A9UDZ9 A0A1B0BB69 Q29IL6 A0A0L0BL70 A0A1A9WL64 A0A1I8NP90 B3NXW2 B4I049 Q9W3W0 B4L526 B4PZ84 B4M701 A0A1W4UW54 B4JLU9 A0A1A9ZQC7 A0A0M4ERE6 A0A067RC69 A0A182YCS3 A0A182P182 A0A182RKY4 U5EG67 A0A182KAD1 A0A2D3C338 A0A1S4FM61 Q16WC0 A0A1Q3FSY0 B0X3V3 A0A182SDK7 A0A182GRZ1 A0A182N0L1 A0A182W5T4 A0A182LY80 A0A0A9Y583 A0A0M9A4X0 J9JUJ3 A0A336MUD6 A0A226DJ08 A0A1D2MQE6 A0A1J1HMG2 A0A0P6CKS2 E9FVS1 T1GKP7 A0A2P8Z3M5 A0A0K2T5U3 T1IMS0
A0A2H1W9C4 F4WYD6 E2BHT3 A0A158NXN7 A0A151WPP2 A0A087ZPY6 D6X1X3 A0A195B0K6 A0A2D3C336 K7IWA0 A0A0L7RJR1 A0A195EVS4 E0W1R2 E2AXW0 A0A154NY96 A0A310SLY8 A0A232FJA6 T1HDJ1 A0A384THG1 A0A3L8E411 A0A195EDX7 A0A151INR2 U4TXE5 A0A1B0DB04 N6U8L1 A0A1I8MW96 A0A3B0KRT7 A0A026WJG3 A0A1A9UDZ9 A0A1B0BB69 Q29IL6 A0A0L0BL70 A0A1A9WL64 A0A1I8NP90 B3NXW2 B4I049 Q9W3W0 B4L526 B4PZ84 B4M701 A0A1W4UW54 B4JLU9 A0A1A9ZQC7 A0A0M4ERE6 A0A067RC69 A0A182YCS3 A0A182P182 A0A182RKY4 U5EG67 A0A182KAD1 A0A2D3C338 A0A1S4FM61 Q16WC0 A0A1Q3FSY0 B0X3V3 A0A182SDK7 A0A182GRZ1 A0A182N0L1 A0A182W5T4 A0A182LY80 A0A0A9Y583 A0A0M9A4X0 J9JUJ3 A0A336MUD6 A0A226DJ08 A0A1D2MQE6 A0A1J1HMG2 A0A0P6CKS2 E9FVS1 T1GKP7 A0A2P8Z3M5 A0A0K2T5U3 T1IMS0
PDB
6E2F
E-value=1.23701e-45,
Score=465
Ontologies
GO
GO:0016021
GO:0005216
GO:0005887
GO:0005262
GO:0006811
GO:0007628
GO:0001964
GO:0043052
GO:0040040
GO:0048060
GO:0009408
GO:0010996
GO:0050884
GO:0007605
GO:0048148
GO:0007608
GO:0006816
GO:0034703
GO:0006812
GO:0005261
GO:0016020
GO:0005515
GO:0005856
GO:0004890
GO:0043564
GO:0006418
GO:0003824
GO:0055085
PANTHER
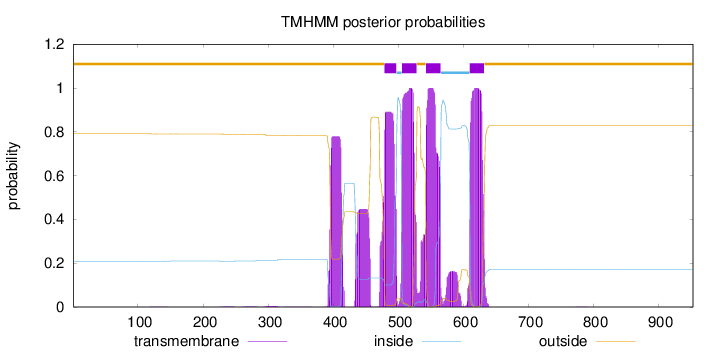
Topology
Length:
953
Number of predicted TMHs:
4
Exp number of AAs in TMHs:
111.95983
Exp number, first 60 AAs:
0.00103
Total prob of N-in:
0.20873
outside
1 - 478
TMhelix
479 - 497
inside
498 - 505
TMhelix
506 - 528
outside
529 - 542
TMhelix
543 - 565
inside
566 - 609
TMhelix
610 - 632
outside
633 - 953
Population Genetic Test Statistics
Pi
36.1316
Theta
17.282916
Tajima's D
-1.89902
CLR
2922.732244
CSRT
0.0209989500524974
Interpretation
Possibly Positive selection
