Gene
KWMTBOMO12031
Pre Gene Modal
BGIBMGA004312
Annotation
PREDICTED:_sodium-dependent_noradrenaline_transporter_[Bombyx_mori]
Full name
Transporter
Location in the cell
PlasmaMembrane Reliability : 4.97
Sequence
CDS
ATGACTTATTGTTTTGTATTTGTTTCAGACCACAAGCCAACGAAGGGATCTGTCGTAACCTTATCGTGCTACAAAAAAGAGCCTAAAGTACAAATAGAGGAAGAGTATTTTTATACCGAAGGCAGCAAACGACTTAGAGCGAACAGCAAGCCTGAAACATCAGATGGCCGAGAGACATGGGGCACCGGGGCAGACTTCCTTCTGTCCATCATTGGTTTCGCGGTTGATCTGGCCAACGTCTGGAGATTTCCGTATCTTTGCTATAGGAATGGCGGTGGGGCCTTCTTGATACCCTACACTCTGATGCTGGTTTTCGGAGCAGTTCCTCTGTTCTACATGGAGTTGATCCTAGGGCAGTATAACCGACAAGGACCTATCACTCTATGGAAAATATGTCCATTGTTTAAAGGTGTTGGATTCTGCGCAGTCATGGTTGCGTTTTATGTTTCCTTCTATTATAATGTAATAATTGGCTGGGCTTTCTACTTCTTGATGTCGTCAGCTCGATCAGAACTGCCCTGGGTGCATTGCGATAATTCTTGGAACACGGAACAATGTTGGGACTCGGCTCGGTTGAATGGTACCAATCGCACGGATGTTAAATACCAAGGACCACTGTCGCATTTCACGCCCGCTTCAGAATTCTTTCATCGAGCAGTCCTGGAAATGCAGCATTCGGAAGGTTTGAATGATCTAGGTTTGCCGAAATGGCAATTGGCTGCTTGTCTCGGTTTGGTCTATGTAACTTTATACCTTTCATTATTCAAAGGCGTTAAAAGCTCCGGAAAAGTGGTATGGATGACAGCAACAATGCCGTACGTTGTCCTGTCGATCCTCCTGGCCCGTGGATTACTTCTGCCGGGAGCAACACGGGGCATCGCCTACTATTTGCAGCCGGAACTTACTAGGCTCAAGGATACGCAGGTTTGGGTGGATGCAGCGGTGCAGATATTTTATTCCGTTGGTGCTGGTTTCGGCGTTCATCTTTCATATGCAAGTTACAACACTTTTCACAACAATTGCTACAGGGATTGCTTGGTTACAACTCTAGTGAATTGTTTCACGTCATTCTTCTCCGGGTTCGTGATCTTCACGTACCTCGGGTTCATGTCGTACAAACAGGGTGTACCCATTTCCTCCGTGGCTACTGAAGGACCAGGATTGGTATTCCAAGTATACCCCGAGGCTGTTGCTACTCTACCGGGGGCGAGCTTATGGGCGATGTTGTTCTTCTTTATGCTGATCATGCTTGGACTGGACTCGGGGATGGGTGGTCTCGAGTGTGTGATCACCGGTCTTTTGGACCAGGCGCGTGCGAGCGGTGCACACTGGCTCCGCCGGGAGCACTTCACGCTGCTGGTCGTCTGTGTGTCCTTCTGTATCGCTTGCATCAATGTAACACCGGGTGGAATTTACATGTTTCATCTTTTGGACACCTACGCTGCCGGCATATCACTGTTGTGCTCGGCGTTGTTCGAAGCCGTTGCTGTCTCGTGGTTTTATGGTCTGAAAAGGTTCTCTGACGATGTTGAAGAAATGCTCGGCTTCAGGCCCGGTATTTACTGGAGGATATGCTGGAAATTTGTCAGCCCAATATTCATTATCGGAGTAGTAGTTTTCGGCTTATTGTACCAGCAGCCTCTTCAGTATCAGCAATACACGTATCCGCAGTGGGCCGTGGTGTTGGGCTGGGGTCTGGCCTGCTCCTCCATATTAATGATACCTATCGTAGCTATATACAAACTGGCCACAACTCCCGGAACATTTAGACAGCGTCTTGCTTGCTGCATTTCTCCTGAATCCGAACACGAAGCCATCCGAGGTGGGGCTCCCGTCAGTCGCTTCACGTGGCGACATTGGTTATATGTATAA
Protein
MTYCFVFVSDHKPTKGSVVTLSCYKKEPKVQIEEEYFYTEGSKRLRANSKPETSDGRETWGTGADFLLSIIGFAVDLANVWRFPYLCYRNGGGAFLIPYTLMLVFGAVPLFYMELILGQYNRQGPITLWKICPLFKGVGFCAVMVAFYVSFYYNVIIGWAFYFLMSSARSELPWVHCDNSWNTEQCWDSARLNGTNRTDVKYQGPLSHFTPASEFFHRAVLEMQHSEGLNDLGLPKWQLAACLGLVYVTLYLSLFKGVKSSGKVVWMTATMPYVVLSILLARGLLLPGATRGIAYYLQPELTRLKDTQVWVDAAVQIFYSVGAGFGVHLSYASYNTFHNNCYRDCLVTTLVNCFTSFFSGFVIFTYLGFMSYKQGVPISSVATEGPGLVFQVYPEAVATLPGASLWAMLFFFMLIMLGLDSGMGGLECVITGLLDQARASGAHWLRREHFTLLVVCVSFCIACINVTPGGIYMFHLLDTYAAGISLLCSALFEAVAVSWFYGLKRFSDDVEEMLGFRPGIYWRICWKFVSPIFIIGVVVFGLLYQQPLQYQQYTYPQWAVVLGWGLACSSILMIPIVAIYKLATTPGTFRQRLACCISPESEHEAIRGGAPVSRFTWRHWLYV
Summary
Similarity
Belongs to the sodium:neurotransmitter symporter (SNF) (TC 2.A.22) family.
Feature
chain Transporter
Uniprot
A0A2A4JW50
Q95VZ4
A0A0G3VI17
A0A194QBN1
Q4F9L7
Q4F9L6
+ More
A0A2H1W3A0 A0A194QQ95 A0A3S2NWE8 A0A212EYB8 H9J473 A0A139WF45 A0A1Y1N0B0 E0VII8 A0A1B6D9T1 A0A067RBN9 A0A224X8S8 A0A023F4G2 A0A2S2RAM0 J9K6Y9 A0A2R5LNN7 A0A147BMK7 T1I6U4 A0A0A9YIP2 U5TBR5 T1J4W0 A0A087T891 U5TBH5 U5T862 B7PMD1 A0A0P4VPR8 A0A0K2USJ4 N6TG93 A0A1S3KVY3 A0A3Q3F619 H2TS08 A0A3P8SMA2 A0A3P8X986 A0A3Q1H5J6 A0A3B4D3D5 A0A3Q1FP96 A0A2U9BD53 A0A3B5AC26 A0A3B4XWP2 W5MR30 W5MR40 G3PWI5 W5KZP3 A0A3P9NHC8 A0A2D0Q645 M4A086 A0A3Q3JGI9 A0A3B3C9X3 A0A3P8XDM8 A0A3B5M1M2 A0A087YE66 A0A3B3X744 A0A146WF78 A0A346RQW8 A0A3P8VBY7 A0A3Q2ED20 A0A1W5ARW5 A0A3B5QMW5 A0A1W5AZG2 A0A3P8V979 A0A1S3IWS7 A0A1L8GE72 A0A3B4C769 F1R3H6 A0A1S3IUX5 W5MJG9 Q90ZV1 A0A2D0S039 A0A3P8XMW3 O42482 R7TSR3 A0A1V4K1V4 A0A3N0XEX3 A0A3B3Q7S6 A0A3Q3B0W7 A0A3B4BBD3 A0A3L8SFH6 A0A3B3R8U0 A0A0Q3TJY1 Q0PKT4 A0A218UYJ4 G1MYP0 A0A2Y9DSN1 H2TS03 A0A315W074 G3WUU1 A0A1A7XVR8 A0A1L8FS41 K7GFU5 A0A0P7VIU1 A0A2I0M7Y9 F7FWA1 H0ZAC2 A0A3P9JNY7
A0A2H1W3A0 A0A194QQ95 A0A3S2NWE8 A0A212EYB8 H9J473 A0A139WF45 A0A1Y1N0B0 E0VII8 A0A1B6D9T1 A0A067RBN9 A0A224X8S8 A0A023F4G2 A0A2S2RAM0 J9K6Y9 A0A2R5LNN7 A0A147BMK7 T1I6U4 A0A0A9YIP2 U5TBR5 T1J4W0 A0A087T891 U5TBH5 U5T862 B7PMD1 A0A0P4VPR8 A0A0K2USJ4 N6TG93 A0A1S3KVY3 A0A3Q3F619 H2TS08 A0A3P8SMA2 A0A3P8X986 A0A3Q1H5J6 A0A3B4D3D5 A0A3Q1FP96 A0A2U9BD53 A0A3B5AC26 A0A3B4XWP2 W5MR30 W5MR40 G3PWI5 W5KZP3 A0A3P9NHC8 A0A2D0Q645 M4A086 A0A3Q3JGI9 A0A3B3C9X3 A0A3P8XDM8 A0A3B5M1M2 A0A087YE66 A0A3B3X744 A0A146WF78 A0A346RQW8 A0A3P8VBY7 A0A3Q2ED20 A0A1W5ARW5 A0A3B5QMW5 A0A1W5AZG2 A0A3P8V979 A0A1S3IWS7 A0A1L8GE72 A0A3B4C769 F1R3H6 A0A1S3IUX5 W5MJG9 Q90ZV1 A0A2D0S039 A0A3P8XMW3 O42482 R7TSR3 A0A1V4K1V4 A0A3N0XEX3 A0A3B3Q7S6 A0A3Q3B0W7 A0A3B4BBD3 A0A3L8SFH6 A0A3B3R8U0 A0A0Q3TJY1 Q0PKT4 A0A218UYJ4 G1MYP0 A0A2Y9DSN1 H2TS03 A0A315W074 G3WUU1 A0A1A7XVR8 A0A1L8FS41 K7GFU5 A0A0P7VIU1 A0A2I0M7Y9 F7FWA1 H0ZAC2 A0A3P9JNY7
Pubmed
11804806
26354079
15986385
22118469
19121390
18362917
+ More
19820115 28004739 20566863 24845553 25474469 29652888 25401762 17142674 23537049 21551351 25069045 25329095 23542700 29451363 30120603 24487278 27762356 23594743 11231083 9092590 23254933 29240929 25463417 30282656 20838655 29703783 21709235 17381049 23371554 17495919 20360741 17554307
19820115 28004739 20566863 24845553 25474469 29652888 25401762 17142674 23537049 21551351 25069045 25329095 23542700 29451363 30120603 24487278 27762356 23594743 11231083 9092590 23254933 29240929 25463417 30282656 20838655 29703783 21709235 17381049 23371554 17495919 20360741 17554307
EMBL
NWSH01000515
PCG75918.1
AF388173
AAL09578.2
KP657645
AKL78870.1
+ More
KQ459460 KPJ00846.1 DQ097796 AAZ08592.2 DQ097797 AAZ08593.2 ODYU01005982 SOQ47426.1 KQ461181 KPJ07672.1 RSAL01000132 RVE46344.1 AGBW02011555 OWR46493.1 BABH01032501 KQ971354 KYB26417.1 GEZM01016170 JAV91351.1 DS235200 EEB13194.1 GEDC01028669 GEDC01014839 JAS08629.1 JAS22459.1 KK852611 KDR20283.1 GFTR01007659 JAW08767.1 GBBI01002650 JAC16062.1 GGMS01017893 MBY87096.1 ABLF02030771 GGLE01006912 MBY11038.1 GEGO01003692 JAR91712.1 ACPB03005112 GBHO01014204 JAG29400.1 KF321729 AGY96957.1 JH431850 KK113908 KFM61330.1 KF321730 AGY96958.1 KF321731 AGY96959.1 ABJB010134719 ABJB010470067 ABJB010499826 ABJB010927746 ABJB011106229 DS746704 EEC07753.1 GDRN01108184 JAI57300.1 HACA01023888 CDW41249.1 APGK01028545 KB740694 ENN79419.1 CP026247 AWP01893.1 AHAT01020181 AHAT01020182 AHAT01020183 AYCK01004742 GCES01057633 GCES01013368 JAR28690.1 MH085049 AXS75745.1 CM004473 OCT82188.1 BX663609 AHAT01020906 AF318177 AAK52449.1 U72877 AAB67676.1 AMQN01000284 KB308724 ELT96928.1 LSYS01005191 OPJ77867.1 RJVU01077109 ROI15904.1 QUSF01000023 RLW01230.1 LMAW01002501 KQK80910.1 DQ813341 ABG91820.1 MUZQ01000095 OWK58581.1 NHOQ01000682 PWA29135.1 AEFK01053212 AEFK01053213 AEFK01053214 AEFK01053215 AEFK01053216 AEFK01053217 AEFK01053218 AEFK01053219 HADW01020475 HADX01008531 SBP21875.1 CM004477 OCT74385.1 AGCU01072366 AGCU01072367 AGCU01072368 AGCU01072369 AGCU01072370 JARO02001429 KPP75498.1 AKCR02000031 PKK25798.1 ABQF01035888 ABQF01035889 ABQF01035890 ABQF01035891
KQ459460 KPJ00846.1 DQ097796 AAZ08592.2 DQ097797 AAZ08593.2 ODYU01005982 SOQ47426.1 KQ461181 KPJ07672.1 RSAL01000132 RVE46344.1 AGBW02011555 OWR46493.1 BABH01032501 KQ971354 KYB26417.1 GEZM01016170 JAV91351.1 DS235200 EEB13194.1 GEDC01028669 GEDC01014839 JAS08629.1 JAS22459.1 KK852611 KDR20283.1 GFTR01007659 JAW08767.1 GBBI01002650 JAC16062.1 GGMS01017893 MBY87096.1 ABLF02030771 GGLE01006912 MBY11038.1 GEGO01003692 JAR91712.1 ACPB03005112 GBHO01014204 JAG29400.1 KF321729 AGY96957.1 JH431850 KK113908 KFM61330.1 KF321730 AGY96958.1 KF321731 AGY96959.1 ABJB010134719 ABJB010470067 ABJB010499826 ABJB010927746 ABJB011106229 DS746704 EEC07753.1 GDRN01108184 JAI57300.1 HACA01023888 CDW41249.1 APGK01028545 KB740694 ENN79419.1 CP026247 AWP01893.1 AHAT01020181 AHAT01020182 AHAT01020183 AYCK01004742 GCES01057633 GCES01013368 JAR28690.1 MH085049 AXS75745.1 CM004473 OCT82188.1 BX663609 AHAT01020906 AF318177 AAK52449.1 U72877 AAB67676.1 AMQN01000284 KB308724 ELT96928.1 LSYS01005191 OPJ77867.1 RJVU01077109 ROI15904.1 QUSF01000023 RLW01230.1 LMAW01002501 KQK80910.1 DQ813341 ABG91820.1 MUZQ01000095 OWK58581.1 NHOQ01000682 PWA29135.1 AEFK01053212 AEFK01053213 AEFK01053214 AEFK01053215 AEFK01053216 AEFK01053217 AEFK01053218 AEFK01053219 HADW01020475 HADX01008531 SBP21875.1 CM004477 OCT74385.1 AGCU01072366 AGCU01072367 AGCU01072368 AGCU01072369 AGCU01072370 JARO02001429 KPP75498.1 AKCR02000031 PKK25798.1 ABQF01035888 ABQF01035889 ABQF01035890 ABQF01035891
Proteomes
UP000218220
UP000053268
UP000053240
UP000283053
UP000007151
UP000005204
+ More
UP000007266 UP000009046 UP000027135 UP000007819 UP000015103 UP000054359 UP000001555 UP000019118 UP000087266 UP000261660 UP000005226 UP000265080 UP000265140 UP000265040 UP000261440 UP000257200 UP000246464 UP000261400 UP000261360 UP000018468 UP000007635 UP000018467 UP000242638 UP000221080 UP000002852 UP000261600 UP000261560 UP000261380 UP000028760 UP000261480 UP000265000 UP000265120 UP000265020 UP000192224 UP000085678 UP000186698 UP000000437 UP000014760 UP000190648 UP000261540 UP000264800 UP000261520 UP000276834 UP000051836 UP000197619 UP000001645 UP000248480 UP000007648 UP000007267 UP000034805 UP000053872 UP000002280 UP000007754 UP000265200
UP000007266 UP000009046 UP000027135 UP000007819 UP000015103 UP000054359 UP000001555 UP000019118 UP000087266 UP000261660 UP000005226 UP000265080 UP000265140 UP000265040 UP000261440 UP000257200 UP000246464 UP000261400 UP000261360 UP000018468 UP000007635 UP000018467 UP000242638 UP000221080 UP000002852 UP000261600 UP000261560 UP000261380 UP000028760 UP000261480 UP000265000 UP000265120 UP000265020 UP000192224 UP000085678 UP000186698 UP000000437 UP000014760 UP000190648 UP000261540 UP000264800 UP000261520 UP000276834 UP000051836 UP000197619 UP000001645 UP000248480 UP000007648 UP000007267 UP000034805 UP000053872 UP000002280 UP000007754 UP000265200
Pfam
Interpro
IPR037272
SNS_sf
+ More
IPR000175 Na/ntran_symport
IPR015153 EF-hand_dom_typ1
IPR015154 EF-hand_dom_typ2
IPR011992 EF-hand-dom_pair
IPR000433 Znf_ZZ
IPR003146 M14A_act_pep
IPR036990 M14A-like_propep
IPR000834 Peptidase_M14
IPR034223 IGCP-like_CPD
IPR002123 Plipid/glycerol_acylTrfase
IPR002048 EF_hand_dom
IPR018247 EF_Hand_1_Ca_BS
IPR002436 Na/ntran_symport_dopamine
IPR002435 Na/ntran_symport_noradrenaline
IPR000175 Na/ntran_symport
IPR015153 EF-hand_dom_typ1
IPR015154 EF-hand_dom_typ2
IPR011992 EF-hand-dom_pair
IPR000433 Znf_ZZ
IPR003146 M14A_act_pep
IPR036990 M14A-like_propep
IPR000834 Peptidase_M14
IPR034223 IGCP-like_CPD
IPR002123 Plipid/glycerol_acylTrfase
IPR002048 EF_hand_dom
IPR018247 EF_Hand_1_Ca_BS
IPR002436 Na/ntran_symport_dopamine
IPR002435 Na/ntran_symport_noradrenaline
Gene 3D
ProteinModelPortal
A0A2A4JW50
Q95VZ4
A0A0G3VI17
A0A194QBN1
Q4F9L7
Q4F9L6
+ More
A0A2H1W3A0 A0A194QQ95 A0A3S2NWE8 A0A212EYB8 H9J473 A0A139WF45 A0A1Y1N0B0 E0VII8 A0A1B6D9T1 A0A067RBN9 A0A224X8S8 A0A023F4G2 A0A2S2RAM0 J9K6Y9 A0A2R5LNN7 A0A147BMK7 T1I6U4 A0A0A9YIP2 U5TBR5 T1J4W0 A0A087T891 U5TBH5 U5T862 B7PMD1 A0A0P4VPR8 A0A0K2USJ4 N6TG93 A0A1S3KVY3 A0A3Q3F619 H2TS08 A0A3P8SMA2 A0A3P8X986 A0A3Q1H5J6 A0A3B4D3D5 A0A3Q1FP96 A0A2U9BD53 A0A3B5AC26 A0A3B4XWP2 W5MR30 W5MR40 G3PWI5 W5KZP3 A0A3P9NHC8 A0A2D0Q645 M4A086 A0A3Q3JGI9 A0A3B3C9X3 A0A3P8XDM8 A0A3B5M1M2 A0A087YE66 A0A3B3X744 A0A146WF78 A0A346RQW8 A0A3P8VBY7 A0A3Q2ED20 A0A1W5ARW5 A0A3B5QMW5 A0A1W5AZG2 A0A3P8V979 A0A1S3IWS7 A0A1L8GE72 A0A3B4C769 F1R3H6 A0A1S3IUX5 W5MJG9 Q90ZV1 A0A2D0S039 A0A3P8XMW3 O42482 R7TSR3 A0A1V4K1V4 A0A3N0XEX3 A0A3B3Q7S6 A0A3Q3B0W7 A0A3B4BBD3 A0A3L8SFH6 A0A3B3R8U0 A0A0Q3TJY1 Q0PKT4 A0A218UYJ4 G1MYP0 A0A2Y9DSN1 H2TS03 A0A315W074 G3WUU1 A0A1A7XVR8 A0A1L8FS41 K7GFU5 A0A0P7VIU1 A0A2I0M7Y9 F7FWA1 H0ZAC2 A0A3P9JNY7
A0A2H1W3A0 A0A194QQ95 A0A3S2NWE8 A0A212EYB8 H9J473 A0A139WF45 A0A1Y1N0B0 E0VII8 A0A1B6D9T1 A0A067RBN9 A0A224X8S8 A0A023F4G2 A0A2S2RAM0 J9K6Y9 A0A2R5LNN7 A0A147BMK7 T1I6U4 A0A0A9YIP2 U5TBR5 T1J4W0 A0A087T891 U5TBH5 U5T862 B7PMD1 A0A0P4VPR8 A0A0K2USJ4 N6TG93 A0A1S3KVY3 A0A3Q3F619 H2TS08 A0A3P8SMA2 A0A3P8X986 A0A3Q1H5J6 A0A3B4D3D5 A0A3Q1FP96 A0A2U9BD53 A0A3B5AC26 A0A3B4XWP2 W5MR30 W5MR40 G3PWI5 W5KZP3 A0A3P9NHC8 A0A2D0Q645 M4A086 A0A3Q3JGI9 A0A3B3C9X3 A0A3P8XDM8 A0A3B5M1M2 A0A087YE66 A0A3B3X744 A0A146WF78 A0A346RQW8 A0A3P8VBY7 A0A3Q2ED20 A0A1W5ARW5 A0A3B5QMW5 A0A1W5AZG2 A0A3P8V979 A0A1S3IWS7 A0A1L8GE72 A0A3B4C769 F1R3H6 A0A1S3IUX5 W5MJG9 Q90ZV1 A0A2D0S039 A0A3P8XMW3 O42482 R7TSR3 A0A1V4K1V4 A0A3N0XEX3 A0A3B3Q7S6 A0A3Q3B0W7 A0A3B4BBD3 A0A3L8SFH6 A0A3B3R8U0 A0A0Q3TJY1 Q0PKT4 A0A218UYJ4 G1MYP0 A0A2Y9DSN1 H2TS03 A0A315W074 G3WUU1 A0A1A7XVR8 A0A1L8FS41 K7GFU5 A0A0P7VIU1 A0A2I0M7Y9 F7FWA1 H0ZAC2 A0A3P9JNY7
PDB
4XPA
E-value=9.02863e-160,
Score=1448
Ontologies
GO
GO:0005328
GO:0016021
GO:0008270
GO:0015293
GO:0005330
GO:0035240
GO:0009986
GO:0030424
GO:0043005
GO:0005886
GO:0004181
GO:0005509
GO:0016746
GO:0005887
GO:0032809
GO:0051583
GO:0042734
GO:0015874
GO:0005334
GO:0048265
GO:0008504
GO:0098810
GO:0003779
GO:0098793
GO:0042053
GO:0007626
GO:0043025
GO:0042416
GO:0042220
GO:0021984
GO:0007595
GO:0060134
GO:0047485
GO:0040018
GO:0016600
GO:0001504
GO:0042420
GO:0007608
GO:0006836
GO:0022891
GO:0005856
GO:0004890
GO:0005216
GO:0043564
GO:0006418
GO:0003824
PANTHER
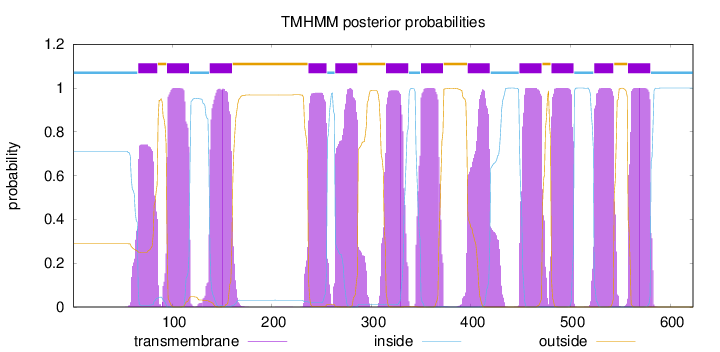
Topology
Length:
623
Number of predicted TMHs:
12
Exp number of AAs in TMHs:
260.59724
Exp number, first 60 AAs:
0.36023
Total prob of N-in:
0.71133
inside
1 - 65
TMhelix
66 - 85
outside
86 - 94
TMhelix
95 - 117
inside
118 - 137
TMhelix
138 - 160
outside
161 - 236
TMhelix
237 - 255
inside
256 - 263
TMhelix
264 - 286
outside
287 - 314
TMhelix
315 - 337
inside
338 - 349
TMhelix
350 - 372
outside
373 - 396
TMhelix
397 - 419
inside
420 - 448
TMhelix
449 - 471
outside
472 - 480
TMhelix
481 - 503
inside
504 - 523
TMhelix
524 - 543
outside
544 - 557
TMhelix
558 - 580
inside
581 - 623
Population Genetic Test Statistics
Pi
233.908139
Theta
191.023341
Tajima's D
0.9982
CLR
0.456477
CSRT
0.654317284135793
Interpretation
Uncertain
