Gene
KWMTBOMO12030
Pre Gene Modal
BGIBMGA004312
Annotation
PREDICTED:_dystrobrevin_beta-like_isoform_X1_[Bombyx_mori]
Full name
Dystrobrevin
+ More
Transporter
Transporter
Location in the cell
Nuclear Reliability : 4.046
Sequence
CDS
ATGTCTTTGGATCGTGAAAGTATAGGAGGTATACCTCCAGTCACCAATGTTGTTGCAGCCGCAGATCCTCGCTCACAACTTCTACAAGAAATGCGAGAACAAAACTTTGATCTAATCAGATTTGCATCATACAGAACAGCTTGTAAACTGCGCTTTGTACAAAAGAAGTGCAACTTACATGTCATAGACATATGGAATGTGATAGAAGCATTTCGTGAGAACGCACTCAACACATTAGAGCCAACGGCCTGTGTAAACGTCACAAGATTAGAAACTTTAGTGTCATCTCTATATCATAATCTGAATAAACGCTTACCGCCTGCACACCAAGTTTCAGTAGAAGCATGCTCAGCGTTACTCTTAAACTGGCTATTGTCAGCATACAGTACTGGTGAGAATGTGGGAAAAATCAGAGTATTCTCAATTAAAGTGGCATTAGCTACAATGTGCGCCGGTAAACTAATGGATAAACTACGATATATATTTTCGCAATTATCAGATGGTAATGGTCATTTATTGATGAAGAGACTGTCTGACTATCTACGTGAAGTACTGGCCTTGCCTGCAGCTGTGTATGAGTCACCATCGTTTAGTTATAATGATACATTGGCACTAACTATTTTTAATCAGAATGGAAAGATAACAGTGAATGACTTCCTAGACACATTAATGTCTGATCCTGGTCCTCCATGCCTGGTCTGGCTACCATTATTGCATCGCCTTGCCAGTGTTGAAAATGTGGTGCACCCGATGTCGTGCAGCGCGTGCCGGCGCGGCTCGCTCACCGGCTTCCGGTACCGCTGCACGCGCTGCGCCAGCTGCACGCTCTGCCAGGACTGCTTCTGGCGGGGCCGCCCGGCGCCGCCGCACGCCGACGACCACGAGGTCAAGGAGTACGCCTCATACAAATCGCCGTCGAAGCAGATCGGCGCTACGTTACGGAAGTCGTTCCGCTGCGTTCCCGAACGGGCCCGAGCCCAGCTGCCGCGGTACCCCGACCAACCGGAGAGGACGCTCAACCTCAGCCATATTGTGCCTCCGTCCCCAGTGCCGGCCCACAACGGCTTCCCGGAGTACGTGGGTGGCTACGTCAACACGGGCTCTTTGGATTCGCGTTCGTCCCGATCCACACATCGCAGTATAGCAGAGCATCTATCGCGTGGAGTGGACGACGAGCACCGCCTCATAGCGCGGTACGCGGCTAGACTGGCCCAGGAAAACCGGACTATGCCTCGTGCTGGAAGATCAGCCTCCTTGACCCCTGAGTTGGGTCGTTCGTCGTCCGAAGGTTCGGAAGTGGACGCGGCTCGACAACAGCGCGAGCTCATCTCACAGCTGGAGGCTAAGAATCGAGAAATTATGAGGGAAATAGCGAGACTCAGACGCCAACAGGAAGCCGAAGCCGGTGCTGGTTCGACGTCGGCCCCGCCGCTGGTGTCGGAGCTGAGAGCCCTCAGACAACGGAAGGACGAACTCGAAGGACATCTCTCCTCGCTTCAGGACTCCAGGAAACACCTAATGCAGCAGCTCGAGGGACTCATGAGGATGCTGAAGACACAACAATCATCACCGCGTTCGACGCCCAATTCGTCTCCTAGATCCACCAAAAGTCCACCTTTGCCGGGAGCTCAAACTCAACCATCTGCACCGGAAAGAGACACGAGCGGCCGCGGCAATGTGAGAAGCGCGCCGCAAACGCCATCTCTCGGCGAGAGAGAGCGGATCGACATGACGCACAGTCTCGGTGCAATGGAGAGCATGTCCAATGAAACGAGATGTTTCCCTCAGAACCAAAGGCCTACGACAATGGGCATAGACAATCGTAGTCTAAGGAGCGACCTTCTGTACGCTGCGGATTCTGTGACGAACGCCATGTCCACTCTGGTCAGAGAACTCAATTCAGAGGGCTCAGACACCGAAGATACGGTTAAAGGCGATTCGAGAAAGCAAAGAGATTTTGAAGAGGACGACGATAGCGACGAGCCAATGCCTAGACCTCAACCGCCGAGACACTATCTACACGATACTAATGTACAAGATACGCCTCCTCCGCTGCCGGCTCGCACGGGACATTACTATCCTCGGACATAA
Protein
MSLDRESIGGIPPVTNVVAAADPRSQLLQEMREQNFDLIRFASYRTACKLRFVQKKCNLHVIDIWNVIEAFRENALNTLEPTACVNVTRLETLVSSLYHNLNKRLPPAHQVSVEACSALLLNWLLSAYSTGENVGKIRVFSIKVALATMCAGKLMDKLRYIFSQLSDGNGHLLMKRLSDYLREVLALPAAVYESPSFSYNDTLALTIFNQNGKITVNDFLDTLMSDPGPPCLVWLPLLHRLASVENVVHPMSCSACRRGSLTGFRYRCTRCASCTLCQDCFWRGRPAPPHADDHEVKEYASYKSPSKQIGATLRKSFRCVPERARAQLPRYPDQPERTLNLSHIVPPSPVPAHNGFPEYVGGYVNTGSLDSRSSRSTHRSIAEHLSRGVDDEHRLIARYAARLAQENRTMPRAGRSASLTPELGRSSSEGSEVDAARQQRELISQLEAKNREIMREIARLRRQQEAEAGAGSTSAPPLVSELRALRQRKDELEGHLSSLQDSRKHLMQQLEGLMRMLKTQQSSPRSTPNSSPRSTKSPPLPGAQTQPSAPERDTSGRGNVRSAPQTPSLGERERIDMTHSLGAMESMSNETRCFPQNQRPTTMGIDNRSLRSDLLYAADSVTNAMSTLVRELNSEGSDTEDTVKGDSRKQRDFEEDDDSDEPMPRPQPPRHYLHDTNVQDTPPPLPARTGHYYPRT
Summary
Similarity
Belongs to the dystrophin family. Dystrobrevin subfamily.
Belongs to the sodium:neurotransmitter symporter (SNF) (TC 2.A.22) family.
Belongs to the sodium:neurotransmitter symporter (SNF) (TC 2.A.22) family.
Uniprot
A0A2W1BP67
A0A194QQ95
A0A194QBN1
H9J473
A0A3S2NFN1
A0A2A4JV55
+ More
A0A067R1F0 A0A2J7RFY3 A0A1B6C2U4 A0A139WH15 A0A139WH50 A0A154P0D3 A0A1Y1LGE0 U4UH02 N6T3D0 A0A1Y1L916 A0A1L8E5R4 K7J5Z9 A0A2A3EPY3 A0A088AKV2 A0A0L7RGD2 A0A026WJC5 A0A1B0GJT1 E2B2N1 J9JV60 E2A3B0 A0A195BL88 A0A195FG05 A0A158NXL6 A0A195ECJ2 A0A151WKN0 F4WQ21 A0A1B6ELE9 A0A0P4VWR2 A0A2H8TH07 A0A084WT44 A0A2S2R3R7 A0A310S7L1 A0A224XD34 A0A0V0G6N6 A0A023F4P2 A0A0Q9VY08 A0A0Q9XFS2 A0A0Q9W5R1 A0A0Q9XI89 A0A0Q9XGE2 A0A0Q9VYL6 A0A0Q9X7P9 B4KR83 A0A182NST0 B4J8V2 B4MFG9 A0A0Q9XK09 A0A182XKB8 A0A182R1C2 A0A182IYX1 A0A034W609 A0A034W5M3 A0A182PCW3 A0A182HJM5 A0A182MRM3 A0A182RH09 A0A0M4EGD2 A0A1B0G459 A0A182VN27 A0A034W1Y4 A0A0K8WGW6 A0A182KG95 A0A182KWT3 A0A0A1X4D6 A0A182TZI7 B0W4E4 A0A3B0JMY2 A0A0K8UP81 A0A1A9V1N9 A0A0P8XLW0 B3MEM7 A0A1A9XUR5 A0A0R3NU80 A0A0A9Y3K0 A0A146KJV6 A0A0A9Y214 A0A1I8PCL7 N6W4T4 A0A1I8PCC6 A0A1I8PCI4 A0A1I8PCK9 A0A0P8ZLL8 A0A182VRX0 Q17AA2 A0A1I8PCL2 A0A1I8PCD2 A0A0P8Y930 W5JUP7 A0A0R3NPH1 N6V8M5 A0A0R3NV40 B4GGH9 A0A1I8PCJ3 A0A1I8PCC3
A0A067R1F0 A0A2J7RFY3 A0A1B6C2U4 A0A139WH15 A0A139WH50 A0A154P0D3 A0A1Y1LGE0 U4UH02 N6T3D0 A0A1Y1L916 A0A1L8E5R4 K7J5Z9 A0A2A3EPY3 A0A088AKV2 A0A0L7RGD2 A0A026WJC5 A0A1B0GJT1 E2B2N1 J9JV60 E2A3B0 A0A195BL88 A0A195FG05 A0A158NXL6 A0A195ECJ2 A0A151WKN0 F4WQ21 A0A1B6ELE9 A0A0P4VWR2 A0A2H8TH07 A0A084WT44 A0A2S2R3R7 A0A310S7L1 A0A224XD34 A0A0V0G6N6 A0A023F4P2 A0A0Q9VY08 A0A0Q9XFS2 A0A0Q9W5R1 A0A0Q9XI89 A0A0Q9XGE2 A0A0Q9VYL6 A0A0Q9X7P9 B4KR83 A0A182NST0 B4J8V2 B4MFG9 A0A0Q9XK09 A0A182XKB8 A0A182R1C2 A0A182IYX1 A0A034W609 A0A034W5M3 A0A182PCW3 A0A182HJM5 A0A182MRM3 A0A182RH09 A0A0M4EGD2 A0A1B0G459 A0A182VN27 A0A034W1Y4 A0A0K8WGW6 A0A182KG95 A0A182KWT3 A0A0A1X4D6 A0A182TZI7 B0W4E4 A0A3B0JMY2 A0A0K8UP81 A0A1A9V1N9 A0A0P8XLW0 B3MEM7 A0A1A9XUR5 A0A0R3NU80 A0A0A9Y3K0 A0A146KJV6 A0A0A9Y214 A0A1I8PCL7 N6W4T4 A0A1I8PCC6 A0A1I8PCI4 A0A1I8PCK9 A0A0P8ZLL8 A0A182VRX0 Q17AA2 A0A1I8PCL2 A0A1I8PCD2 A0A0P8Y930 W5JUP7 A0A0R3NPH1 N6V8M5 A0A0R3NV40 B4GGH9 A0A1I8PCJ3 A0A1I8PCC3
Pubmed
EMBL
KZ150034
PZC74656.1
KQ461181
KPJ07672.1
KQ459460
KPJ00846.1
+ More
BABH01032501 RSAL01000132 RVE46343.1 NWSH01000515 PCG75917.1 KK852816 KDR15790.1 NEVH01004409 PNF39730.1 GEDC01029719 GEDC01012440 JAS07579.1 JAS24858.1 KQ971343 KYB27288.1 KYB27292.1 KQ434791 KZC05379.1 GEZM01062060 GEZM01062054 JAV70087.1 KB632319 ERL92307.1 APGK01045625 KB741037 ENN74644.1 GEZM01062059 GEZM01062057 JAV70084.1 GFDF01000100 JAV13984.1 KZ288195 PBC33788.1 KQ414599 KOC69786.1 KK107168 QOIP01000001 EZA56132.1 RLU27519.1 AJWK01022211 AJWK01022212 AJWK01022213 AJWK01022214 AJWK01022215 GL445190 EFN90053.1 ABLF02029450 GL436354 EFN72082.1 KQ976441 KYM86432.1 KQ981610 KYN39323.1 ADTU01003143 KQ979074 KYN22933.1 KQ983001 KYQ48439.1 GL888262 EGI63719.1 GECZ01031067 JAS38702.1 GDKW01002557 JAI54038.1 GFXV01001579 MBW13384.1 ATLV01026830 ATLV01026831 KE525419 KFB53388.1 GGMS01015435 MBY84638.1 KQ765323 OAD53979.1 GFTR01007532 JAW08894.1 GECL01002704 JAP03420.1 GBBI01002759 JAC15953.1 CH940667 KRF77624.1 CH933808 KRG04020.1 KRF77623.1 KRG04017.1 KRG04018.1 KRF77622.1 KRG04021.1 EDW08270.2 CH916367 EDW02392.1 EDW57140.2 KRG04019.1 AXCN02000300 GAKP01009367 JAC49585.1 GAKP01009365 JAC49587.1 APCN01002191 AXCM01009120 CP012524 ALC41447.1 CCAG010005748 GAKP01009366 GAKP01009364 JAC49586.1 GDHF01002199 JAI50115.1 GBXI01008754 JAD05538.1 DS231836 EDS33229.1 OUUW01000001 SPP73991.1 GDHF01024021 JAI28293.1 CH902619 KPU75695.1 EDV35491.1 CM000071 KRT02879.1 GBHO01017916 JAG25688.1 GDHC01021766 JAP96862.1 GBHO01017918 JAG25686.1 ENO01807.1 KPU75693.1 CH477337 EAT43189.1 KPU75690.1 ADMH02000406 ETN66675.1 KRT02882.1 ENO01806.2 KRT02885.1 CH479183 EDW35599.1
BABH01032501 RSAL01000132 RVE46343.1 NWSH01000515 PCG75917.1 KK852816 KDR15790.1 NEVH01004409 PNF39730.1 GEDC01029719 GEDC01012440 JAS07579.1 JAS24858.1 KQ971343 KYB27288.1 KYB27292.1 KQ434791 KZC05379.1 GEZM01062060 GEZM01062054 JAV70087.1 KB632319 ERL92307.1 APGK01045625 KB741037 ENN74644.1 GEZM01062059 GEZM01062057 JAV70084.1 GFDF01000100 JAV13984.1 KZ288195 PBC33788.1 KQ414599 KOC69786.1 KK107168 QOIP01000001 EZA56132.1 RLU27519.1 AJWK01022211 AJWK01022212 AJWK01022213 AJWK01022214 AJWK01022215 GL445190 EFN90053.1 ABLF02029450 GL436354 EFN72082.1 KQ976441 KYM86432.1 KQ981610 KYN39323.1 ADTU01003143 KQ979074 KYN22933.1 KQ983001 KYQ48439.1 GL888262 EGI63719.1 GECZ01031067 JAS38702.1 GDKW01002557 JAI54038.1 GFXV01001579 MBW13384.1 ATLV01026830 ATLV01026831 KE525419 KFB53388.1 GGMS01015435 MBY84638.1 KQ765323 OAD53979.1 GFTR01007532 JAW08894.1 GECL01002704 JAP03420.1 GBBI01002759 JAC15953.1 CH940667 KRF77624.1 CH933808 KRG04020.1 KRF77623.1 KRG04017.1 KRG04018.1 KRF77622.1 KRG04021.1 EDW08270.2 CH916367 EDW02392.1 EDW57140.2 KRG04019.1 AXCN02000300 GAKP01009367 JAC49585.1 GAKP01009365 JAC49587.1 APCN01002191 AXCM01009120 CP012524 ALC41447.1 CCAG010005748 GAKP01009366 GAKP01009364 JAC49586.1 GDHF01002199 JAI50115.1 GBXI01008754 JAD05538.1 DS231836 EDS33229.1 OUUW01000001 SPP73991.1 GDHF01024021 JAI28293.1 CH902619 KPU75695.1 EDV35491.1 CM000071 KRT02879.1 GBHO01017916 JAG25688.1 GDHC01021766 JAP96862.1 GBHO01017918 JAG25686.1 ENO01807.1 KPU75693.1 CH477337 EAT43189.1 KPU75690.1 ADMH02000406 ETN66675.1 KRT02882.1 ENO01806.2 KRT02885.1 CH479183 EDW35599.1
Proteomes
UP000053240
UP000053268
UP000005204
UP000283053
UP000218220
UP000027135
+ More
UP000235965 UP000007266 UP000076502 UP000030742 UP000019118 UP000002358 UP000242457 UP000005203 UP000053825 UP000053097 UP000279307 UP000092461 UP000008237 UP000007819 UP000000311 UP000078540 UP000078541 UP000005205 UP000078492 UP000075809 UP000007755 UP000030765 UP000008792 UP000009192 UP000075884 UP000001070 UP000076407 UP000075886 UP000075880 UP000075885 UP000075840 UP000075883 UP000075900 UP000092553 UP000092444 UP000075903 UP000075881 UP000075882 UP000075902 UP000002320 UP000268350 UP000078200 UP000007801 UP000092443 UP000001819 UP000095300 UP000075920 UP000008820 UP000000673 UP000008744
UP000235965 UP000007266 UP000076502 UP000030742 UP000019118 UP000002358 UP000242457 UP000005203 UP000053825 UP000053097 UP000279307 UP000092461 UP000008237 UP000007819 UP000000311 UP000078540 UP000078541 UP000005205 UP000078492 UP000075809 UP000007755 UP000030765 UP000008792 UP000009192 UP000075884 UP000001070 UP000076407 UP000075886 UP000075880 UP000075885 UP000075840 UP000075883 UP000075900 UP000092553 UP000092444 UP000075903 UP000075881 UP000075882 UP000075902 UP000002320 UP000268350 UP000078200 UP000007801 UP000092443 UP000001819 UP000095300 UP000075920 UP000008820 UP000000673 UP000008744
PRIDE
Pfam
Interpro
Gene 3D
ProteinModelPortal
A0A2W1BP67
A0A194QQ95
A0A194QBN1
H9J473
A0A3S2NFN1
A0A2A4JV55
+ More
A0A067R1F0 A0A2J7RFY3 A0A1B6C2U4 A0A139WH15 A0A139WH50 A0A154P0D3 A0A1Y1LGE0 U4UH02 N6T3D0 A0A1Y1L916 A0A1L8E5R4 K7J5Z9 A0A2A3EPY3 A0A088AKV2 A0A0L7RGD2 A0A026WJC5 A0A1B0GJT1 E2B2N1 J9JV60 E2A3B0 A0A195BL88 A0A195FG05 A0A158NXL6 A0A195ECJ2 A0A151WKN0 F4WQ21 A0A1B6ELE9 A0A0P4VWR2 A0A2H8TH07 A0A084WT44 A0A2S2R3R7 A0A310S7L1 A0A224XD34 A0A0V0G6N6 A0A023F4P2 A0A0Q9VY08 A0A0Q9XFS2 A0A0Q9W5R1 A0A0Q9XI89 A0A0Q9XGE2 A0A0Q9VYL6 A0A0Q9X7P9 B4KR83 A0A182NST0 B4J8V2 B4MFG9 A0A0Q9XK09 A0A182XKB8 A0A182R1C2 A0A182IYX1 A0A034W609 A0A034W5M3 A0A182PCW3 A0A182HJM5 A0A182MRM3 A0A182RH09 A0A0M4EGD2 A0A1B0G459 A0A182VN27 A0A034W1Y4 A0A0K8WGW6 A0A182KG95 A0A182KWT3 A0A0A1X4D6 A0A182TZI7 B0W4E4 A0A3B0JMY2 A0A0K8UP81 A0A1A9V1N9 A0A0P8XLW0 B3MEM7 A0A1A9XUR5 A0A0R3NU80 A0A0A9Y3K0 A0A146KJV6 A0A0A9Y214 A0A1I8PCL7 N6W4T4 A0A1I8PCC6 A0A1I8PCI4 A0A1I8PCK9 A0A0P8ZLL8 A0A182VRX0 Q17AA2 A0A1I8PCL2 A0A1I8PCD2 A0A0P8Y930 W5JUP7 A0A0R3NPH1 N6V8M5 A0A0R3NV40 B4GGH9 A0A1I8PCJ3 A0A1I8PCC3
A0A067R1F0 A0A2J7RFY3 A0A1B6C2U4 A0A139WH15 A0A139WH50 A0A154P0D3 A0A1Y1LGE0 U4UH02 N6T3D0 A0A1Y1L916 A0A1L8E5R4 K7J5Z9 A0A2A3EPY3 A0A088AKV2 A0A0L7RGD2 A0A026WJC5 A0A1B0GJT1 E2B2N1 J9JV60 E2A3B0 A0A195BL88 A0A195FG05 A0A158NXL6 A0A195ECJ2 A0A151WKN0 F4WQ21 A0A1B6ELE9 A0A0P4VWR2 A0A2H8TH07 A0A084WT44 A0A2S2R3R7 A0A310S7L1 A0A224XD34 A0A0V0G6N6 A0A023F4P2 A0A0Q9VY08 A0A0Q9XFS2 A0A0Q9W5R1 A0A0Q9XI89 A0A0Q9XGE2 A0A0Q9VYL6 A0A0Q9X7P9 B4KR83 A0A182NST0 B4J8V2 B4MFG9 A0A0Q9XK09 A0A182XKB8 A0A182R1C2 A0A182IYX1 A0A034W609 A0A034W5M3 A0A182PCW3 A0A182HJM5 A0A182MRM3 A0A182RH09 A0A0M4EGD2 A0A1B0G459 A0A182VN27 A0A034W1Y4 A0A0K8WGW6 A0A182KG95 A0A182KWT3 A0A0A1X4D6 A0A182TZI7 B0W4E4 A0A3B0JMY2 A0A0K8UP81 A0A1A9V1N9 A0A0P8XLW0 B3MEM7 A0A1A9XUR5 A0A0R3NU80 A0A0A9Y3K0 A0A146KJV6 A0A0A9Y214 A0A1I8PCL7 N6W4T4 A0A1I8PCC6 A0A1I8PCI4 A0A1I8PCK9 A0A0P8ZLL8 A0A182VRX0 Q17AA2 A0A1I8PCL2 A0A1I8PCD2 A0A0P8Y930 W5JUP7 A0A0R3NPH1 N6V8M5 A0A0R3NV40 B4GGH9 A0A1I8PCJ3 A0A1I8PCC3
PDB
1EG4
E-value=2.99734e-20,
Score=245
Ontologies
GO
PANTHER
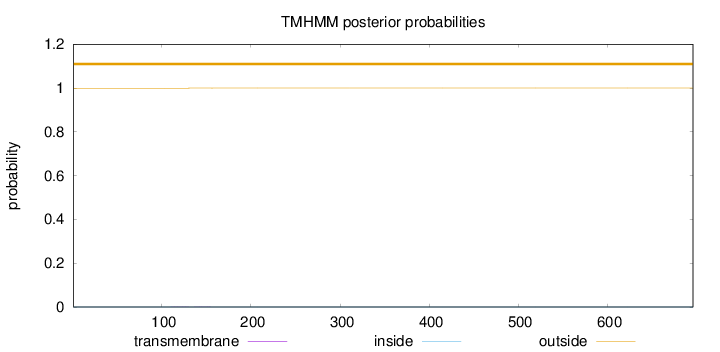
Topology
Subcellular location
Cytoplasm
Length:
696
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00918999999999999
Exp number, first 60 AAs:
0.00175
Total prob of N-in:
0.00051
outside
1 - 696
Population Genetic Test Statistics
Pi
24.495441
Theta
188.384682
Tajima's D
1.243275
CLR
0.423864
CSRT
0.719814009299535
Interpretation
Uncertain
