Gene
KWMTBOMO12029
Pre Gene Modal
BGIBMGA004313
Annotation
PREDICTED:_facilitated_trehalose_transporter_Tret1-like_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.774
Sequence
CDS
ATGGGACCCCGGCTGATGGCCTCCGACACCCCTGCCTCGGAACCCGTTCACGCTGACGTAATATCCTGGATGGCTTCCGTGGTTTATCTGGTTGGTACACCTGCGGTTTTCCTGTTCGGATACATCGTTGACAACTATGGGAGAAAGAAGGCTCTGATGCTGACCTCGTTTTCAATGGCTGCATGTTGGGCTTTAAAGCTGTATTCCACGGAGACCTGGGCTCTAATCACGGCACGTGCGATCATCGGCTTAGGGGTCAGCGGCTCGTATGTCGTCACGCCTCTCTACATCAAGGAAATAAGTGAAGATTCCATCAGGGGGACCCTCGGAAGTCTCGTGGTCTTATCGCAAAACCTAGGCAACCTAGCCGTTTATATATTAGGCGAATACCTGAGCTATTACACGACCCTGTGGATATGTTTGGCAGTGCCGCTTTTGCACATGCTTGTGTTCACGACGATGCCCGACACTCCGTCGTACTTACTGAAAAGTGGTAAAGTTGAGGAAGCTCGCGATGCTCTGGCCTGGCTGCGATGCCGCGATAGCTCCGAAGCCGAAGTGGACAGCGAGCTGCAGAAGCTGTTAGTTGAGCTGGAACAAAGCAAATCTGGAAAATTTGTCAGCGCTGTCAAAACTCTAATATCAGACAGAAATACGTTCCGGGCTTTTCGTATAACCCTCGTGATAACTCTGGCTCGAGAGCTTTGCGGTTGCCTAGCCGTGCTACACTTCGCTTCGCTGATATTCAGCAAGGCCAGCTCGGGCTGGGTGTTGACTGCGAACCAGCAGGCCGTGGTGCTGGGCGTCGTGCAACTGATCGGCTCGTGCACCGCATCGACTCTAGTCGAAAGGACTGGTAGAAAGCCTCTACTCGGCACGACGTGTTTAGTGTCTGGTATATCGATGTGCGCGTTAGGAGCGTGGTTCTACGCGCAGAGCGTCAGCGCCACCGGCGTGTCGTGGCTGCCGATAGCGGCGTTATGTTTGTGCATCTATTGTGATGCGGCCGGACTGCAACCCGTGCCTTTCGTTATCATGACCGAAATGTTCTCTTTCCAGCTCCGCGGTACAGTCACTTCAATAGTGATATCGTTCGCGTGTGCTCTAGTATCGGTTGAGCTCCGCGTGTTCCAACCCCTGGCGGCCTCAATGGGTCTCCATATGGTGTTCTGGCTGTTCTCGGCTGTCTGCATCATCAGCACAGTGTACATAGCTTGCATGGTGCCGGAGACAAAAATGCGAACCCTGGACGAGATCTACACTGAAATAGGTGGCAAGAAACAGAAGGATTGCGAGGCAGACGTCACTAAATTGTAA
Protein
MGPRLMASDTPASEPVHADVISWMASVVYLVGTPAVFLFGYIVDNYGRKKALMLTSFSMAACWALKLYSTETWALITARAIIGLGVSGSYVVTPLYIKEISEDSIRGTLGSLVVLSQNLGNLAVYILGEYLSYYTTLWICLAVPLLHMLVFTTMPDTPSYLLKSGKVEEARDALAWLRCRDSSEAEVDSELQKLLVELEQSKSGKFVSAVKTLISDRNTFRAFRITLVITLARELCGCLAVLHFASLIFSKASSGWVLTANQQAVVLGVVQLIGSCTASTLVERTGRKPLLGTTCLVSGISMCALGAWFYAQSVSATGVSWLPIAALCLCIYCDAAGLQPVPFVIMTEMFSFQLRGTVTSIVISFACALVSVELRVFQPLAASMGLHMVFWLFSAVCIISTVYIACMVPETKMRTLDEIYTEIGGKKQKDCEADVTKL
Summary
Similarity
Belongs to the major facilitator superfamily. Sugar transporter (TC 2.A.1.1) family.
Uniprot
A0A2H1WE18
A0A2A4JHM3
A0A194Q5M1
A0A212FI50
A0A3S2L5Q4
A0A194QQ84
+ More
H9J4J6 A0A194QB83 A0A3S2TIH7 A0A212FAG2 A0A2W1BRA6 A0A2A4J8P9 A0A2W1BQ47 H9JVU3 A0A2A4J7G4 A0A194Q5F6 A0A2A4JII3 A0A212EX46 A0A212FB89 A0A0N1IIG7 A0A2W1BV28 A0A194Q5B9 A0A2H1WQ53 A0A2H1WGI4 A0A3S2TMD3 A0A2Z5U782 A0A0N0PER8 H9J4P6 A0A212F931 A0A0N1IHX2 A0A0N1PHR0 A0A194Q5G2 A0A212F0G4 A0A2H1WR29 A0A0N0PEG1 A0A2A4JKE4 A0A2W1BSA0 A0A2H1WM77 S4PKR1 A0A0L7LE34 A0A2A4JNL8 A0A146LSN9 A0A182JZA6 A0A182NGP0 A0A182QRF9 A0A182XNP0 A0A182P8A2 A0A182UH29 A0A1S4GZI9 Q7PWP0 A0A182MPL4 A0A2A4J9N1 A0A182TXA5 A0A182I799 A0A182LMV4 A0A182VUC8 A0A182UZ66 A0A1B6CY32 A0A2P8XJ31 A0A1B6DME3 A0A2M3Z412 A0A224XJ44 A0A0L7KVL6 A0A194Q5D6 A0A2H1X046 A0A0A8J8K5 A0A2M3ZH26 A0A2M4BLL7 A0A2M3Z428 A0A2M4CQF1 A0A2M4CQC1 W5JMX1 A0A2M4A741 A0A2M4A739 A0A2J7R8L3 A0A182GVZ3 A0A2M4A7E4 A0A2M4BLK9 A0A084W7N7 A0A182RSI8 A0A182JME0 A0A182GL71 A0A182F539 A0A0A9X9R8 Q16SU3 A0A2J7R8M9 A0A1Q3FRY2 A0A146L3L7 B0WPL1 A0A2P8Z801 K7IP31 A0A2P8ZDM8 A0A1Q3FRU2 A0A182YRN8 A0A1B6GYS0 A0A1B6INA2 A0A195FTD3
H9J4J6 A0A194QB83 A0A3S2TIH7 A0A212FAG2 A0A2W1BRA6 A0A2A4J8P9 A0A2W1BQ47 H9JVU3 A0A2A4J7G4 A0A194Q5F6 A0A2A4JII3 A0A212EX46 A0A212FB89 A0A0N1IIG7 A0A2W1BV28 A0A194Q5B9 A0A2H1WQ53 A0A2H1WGI4 A0A3S2TMD3 A0A2Z5U782 A0A0N0PER8 H9J4P6 A0A212F931 A0A0N1IHX2 A0A0N1PHR0 A0A194Q5G2 A0A212F0G4 A0A2H1WR29 A0A0N0PEG1 A0A2A4JKE4 A0A2W1BSA0 A0A2H1WM77 S4PKR1 A0A0L7LE34 A0A2A4JNL8 A0A146LSN9 A0A182JZA6 A0A182NGP0 A0A182QRF9 A0A182XNP0 A0A182P8A2 A0A182UH29 A0A1S4GZI9 Q7PWP0 A0A182MPL4 A0A2A4J9N1 A0A182TXA5 A0A182I799 A0A182LMV4 A0A182VUC8 A0A182UZ66 A0A1B6CY32 A0A2P8XJ31 A0A1B6DME3 A0A2M3Z412 A0A224XJ44 A0A0L7KVL6 A0A194Q5D6 A0A2H1X046 A0A0A8J8K5 A0A2M3ZH26 A0A2M4BLL7 A0A2M3Z428 A0A2M4CQF1 A0A2M4CQC1 W5JMX1 A0A2M4A741 A0A2M4A739 A0A2J7R8L3 A0A182GVZ3 A0A2M4A7E4 A0A2M4BLK9 A0A084W7N7 A0A182RSI8 A0A182JME0 A0A182GL71 A0A182F539 A0A0A9X9R8 Q16SU3 A0A2J7R8M9 A0A1Q3FRY2 A0A146L3L7 B0WPL1 A0A2P8Z801 K7IP31 A0A2P8ZDM8 A0A1Q3FRU2 A0A182YRN8 A0A1B6GYS0 A0A1B6INA2 A0A195FTD3
Pubmed
EMBL
ODYU01008039
SOQ51308.1
NWSH01001423
PCG71276.1
KQ459460
KPJ00847.1
+ More
AGBW02008435 OWR53407.1 RSAL01000132 RVE46342.1 KQ461181 KPJ07673.1 BABH01021836 KPJ00686.1 RSAL01000114 RVE46930.1 AGBW02009482 OWR50710.1 KZ149924 PZC77632.1 NWSH01002620 PCG67830.1 KZ149937 PZC77108.1 BABH01040655 NWSH01002833 PCG67464.1 KPJ00782.1 NWSH01001270 PCG71905.1 AGBW02011864 OWR46004.1 AGBW02009367 OWR50987.1 KQ459838 KPJ19742.1 KZ149954 PZC76506.1 KPJ00738.1 ODYU01010219 SOQ55189.1 ODYU01008186 SOQ51574.1 RSAL01000053 RVE50024.1 AP017499 BBB06788.1 KPJ19739.1 BABH01022059 AGBW02009650 OWR50256.1 KQ460009 KPJ18480.1 KQ460654 KPJ12970.1 KPJ00783.1 AGBW02011078 OWR47246.1 ODYU01010429 SOQ55525.1 KPJ19738.1 PCG71903.1 PZC76505.1 ODYU01009532 SOQ54032.1 GAIX01000896 JAA91664.1 JTDY01001495 KOB73737.1 NWSH01001033 PCG73010.1 GDHC01008360 GDHC01004923 JAQ10269.1 JAQ13706.1 AXCN02002080 AAAB01008984 EAA14882.4 AXCM01000345 NWSH01002244 PCG68837.1 APCN01003374 GEDC01018964 JAS18334.1 PYGN01001954 PSN32011.1 GEDC01010470 JAS26828.1 GGFM01002508 MBW23259.1 GFTR01006588 JAW09838.1 JTDY01005298 KOB67145.1 KPJ00762.1 ODYU01012350 SOQ58622.1 AB901068 BAQ02368.1 GGFM01007009 MBW27760.1 GGFJ01004829 MBW53970.1 GGFM01002530 MBW23281.1 GGFL01003355 MBW67533.1 GGFL01003356 MBW67534.1 ADMH02001096 ETN64119.1 GGFK01003296 MBW36617.1 GGFK01003278 MBW36599.1 NEVH01006721 PNF37174.1 JXUM01091986 KQ563956 KXJ73023.1 GGFK01003378 MBW36699.1 GGFJ01004828 MBW53969.1 ATLV01021268 KE525315 KFB46231.1 JXUM01071371 KQ562660 KXJ75380.1 GBHO01026885 GBRD01008360 JAG16719.1 JAG57461.1 CH477665 EAT37545.1 PNF37175.1 GFDL01004852 JAV30193.1 GDHC01015631 JAQ02998.1 DS232026 EDS32412.1 PYGN01000154 PSN52620.1 PYGN01000088 PSN54598.1 GFDL01004839 JAV30206.1 GECZ01003576 GECZ01002212 JAS66193.1 JAS67557.1 GECU01019308 JAS88398.1 KQ981276 KYN43567.1
AGBW02008435 OWR53407.1 RSAL01000132 RVE46342.1 KQ461181 KPJ07673.1 BABH01021836 KPJ00686.1 RSAL01000114 RVE46930.1 AGBW02009482 OWR50710.1 KZ149924 PZC77632.1 NWSH01002620 PCG67830.1 KZ149937 PZC77108.1 BABH01040655 NWSH01002833 PCG67464.1 KPJ00782.1 NWSH01001270 PCG71905.1 AGBW02011864 OWR46004.1 AGBW02009367 OWR50987.1 KQ459838 KPJ19742.1 KZ149954 PZC76506.1 KPJ00738.1 ODYU01010219 SOQ55189.1 ODYU01008186 SOQ51574.1 RSAL01000053 RVE50024.1 AP017499 BBB06788.1 KPJ19739.1 BABH01022059 AGBW02009650 OWR50256.1 KQ460009 KPJ18480.1 KQ460654 KPJ12970.1 KPJ00783.1 AGBW02011078 OWR47246.1 ODYU01010429 SOQ55525.1 KPJ19738.1 PCG71903.1 PZC76505.1 ODYU01009532 SOQ54032.1 GAIX01000896 JAA91664.1 JTDY01001495 KOB73737.1 NWSH01001033 PCG73010.1 GDHC01008360 GDHC01004923 JAQ10269.1 JAQ13706.1 AXCN02002080 AAAB01008984 EAA14882.4 AXCM01000345 NWSH01002244 PCG68837.1 APCN01003374 GEDC01018964 JAS18334.1 PYGN01001954 PSN32011.1 GEDC01010470 JAS26828.1 GGFM01002508 MBW23259.1 GFTR01006588 JAW09838.1 JTDY01005298 KOB67145.1 KPJ00762.1 ODYU01012350 SOQ58622.1 AB901068 BAQ02368.1 GGFM01007009 MBW27760.1 GGFJ01004829 MBW53970.1 GGFM01002530 MBW23281.1 GGFL01003355 MBW67533.1 GGFL01003356 MBW67534.1 ADMH02001096 ETN64119.1 GGFK01003296 MBW36617.1 GGFK01003278 MBW36599.1 NEVH01006721 PNF37174.1 JXUM01091986 KQ563956 KXJ73023.1 GGFK01003378 MBW36699.1 GGFJ01004828 MBW53969.1 ATLV01021268 KE525315 KFB46231.1 JXUM01071371 KQ562660 KXJ75380.1 GBHO01026885 GBRD01008360 JAG16719.1 JAG57461.1 CH477665 EAT37545.1 PNF37175.1 GFDL01004852 JAV30193.1 GDHC01015631 JAQ02998.1 DS232026 EDS32412.1 PYGN01000154 PSN52620.1 PYGN01000088 PSN54598.1 GFDL01004839 JAV30206.1 GECZ01003576 GECZ01002212 JAS66193.1 JAS67557.1 GECU01019308 JAS88398.1 KQ981276 KYN43567.1
Proteomes
UP000218220
UP000053268
UP000007151
UP000283053
UP000053240
UP000005204
+ More
UP000037510 UP000075881 UP000075884 UP000075886 UP000076407 UP000075885 UP000075902 UP000007062 UP000075883 UP000075840 UP000075882 UP000075920 UP000075903 UP000245037 UP000000673 UP000235965 UP000069940 UP000249989 UP000030765 UP000075900 UP000075880 UP000069272 UP000008820 UP000002320 UP000002358 UP000076408 UP000078541
UP000037510 UP000075881 UP000075884 UP000075886 UP000076407 UP000075885 UP000075902 UP000007062 UP000075883 UP000075840 UP000075882 UP000075920 UP000075903 UP000245037 UP000000673 UP000235965 UP000069940 UP000249989 UP000030765 UP000075900 UP000075880 UP000069272 UP000008820 UP000002320 UP000002358 UP000076408 UP000078541
Interpro
CDD
ProteinModelPortal
A0A2H1WE18
A0A2A4JHM3
A0A194Q5M1
A0A212FI50
A0A3S2L5Q4
A0A194QQ84
+ More
H9J4J6 A0A194QB83 A0A3S2TIH7 A0A212FAG2 A0A2W1BRA6 A0A2A4J8P9 A0A2W1BQ47 H9JVU3 A0A2A4J7G4 A0A194Q5F6 A0A2A4JII3 A0A212EX46 A0A212FB89 A0A0N1IIG7 A0A2W1BV28 A0A194Q5B9 A0A2H1WQ53 A0A2H1WGI4 A0A3S2TMD3 A0A2Z5U782 A0A0N0PER8 H9J4P6 A0A212F931 A0A0N1IHX2 A0A0N1PHR0 A0A194Q5G2 A0A212F0G4 A0A2H1WR29 A0A0N0PEG1 A0A2A4JKE4 A0A2W1BSA0 A0A2H1WM77 S4PKR1 A0A0L7LE34 A0A2A4JNL8 A0A146LSN9 A0A182JZA6 A0A182NGP0 A0A182QRF9 A0A182XNP0 A0A182P8A2 A0A182UH29 A0A1S4GZI9 Q7PWP0 A0A182MPL4 A0A2A4J9N1 A0A182TXA5 A0A182I799 A0A182LMV4 A0A182VUC8 A0A182UZ66 A0A1B6CY32 A0A2P8XJ31 A0A1B6DME3 A0A2M3Z412 A0A224XJ44 A0A0L7KVL6 A0A194Q5D6 A0A2H1X046 A0A0A8J8K5 A0A2M3ZH26 A0A2M4BLL7 A0A2M3Z428 A0A2M4CQF1 A0A2M4CQC1 W5JMX1 A0A2M4A741 A0A2M4A739 A0A2J7R8L3 A0A182GVZ3 A0A2M4A7E4 A0A2M4BLK9 A0A084W7N7 A0A182RSI8 A0A182JME0 A0A182GL71 A0A182F539 A0A0A9X9R8 Q16SU3 A0A2J7R8M9 A0A1Q3FRY2 A0A146L3L7 B0WPL1 A0A2P8Z801 K7IP31 A0A2P8ZDM8 A0A1Q3FRU2 A0A182YRN8 A0A1B6GYS0 A0A1B6INA2 A0A195FTD3
H9J4J6 A0A194QB83 A0A3S2TIH7 A0A212FAG2 A0A2W1BRA6 A0A2A4J8P9 A0A2W1BQ47 H9JVU3 A0A2A4J7G4 A0A194Q5F6 A0A2A4JII3 A0A212EX46 A0A212FB89 A0A0N1IIG7 A0A2W1BV28 A0A194Q5B9 A0A2H1WQ53 A0A2H1WGI4 A0A3S2TMD3 A0A2Z5U782 A0A0N0PER8 H9J4P6 A0A212F931 A0A0N1IHX2 A0A0N1PHR0 A0A194Q5G2 A0A212F0G4 A0A2H1WR29 A0A0N0PEG1 A0A2A4JKE4 A0A2W1BSA0 A0A2H1WM77 S4PKR1 A0A0L7LE34 A0A2A4JNL8 A0A146LSN9 A0A182JZA6 A0A182NGP0 A0A182QRF9 A0A182XNP0 A0A182P8A2 A0A182UH29 A0A1S4GZI9 Q7PWP0 A0A182MPL4 A0A2A4J9N1 A0A182TXA5 A0A182I799 A0A182LMV4 A0A182VUC8 A0A182UZ66 A0A1B6CY32 A0A2P8XJ31 A0A1B6DME3 A0A2M3Z412 A0A224XJ44 A0A0L7KVL6 A0A194Q5D6 A0A2H1X046 A0A0A8J8K5 A0A2M3ZH26 A0A2M4BLL7 A0A2M3Z428 A0A2M4CQF1 A0A2M4CQC1 W5JMX1 A0A2M4A741 A0A2M4A739 A0A2J7R8L3 A0A182GVZ3 A0A2M4A7E4 A0A2M4BLK9 A0A084W7N7 A0A182RSI8 A0A182JME0 A0A182GL71 A0A182F539 A0A0A9X9R8 Q16SU3 A0A2J7R8M9 A0A1Q3FRY2 A0A146L3L7 B0WPL1 A0A2P8Z801 K7IP31 A0A2P8ZDM8 A0A1Q3FRU2 A0A182YRN8 A0A1B6GYS0 A0A1B6INA2 A0A195FTD3
PDB
4LDS
E-value=6.3729e-25,
Score=283
Ontologies
GO
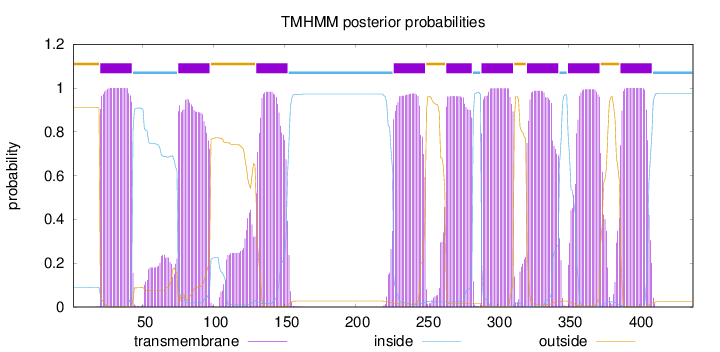
Topology
Length:
438
Number of predicted TMHs:
9
Exp number of AAs in TMHs:
204.04432
Exp number, first 60 AAs:
24.48106
Total prob of N-in:
0.08849
POSSIBLE N-term signal
sequence
outside
1 - 19
TMhelix
20 - 42
inside
43 - 74
TMhelix
75 - 97
outside
98 - 129
TMhelix
130 - 152
inside
153 - 226
TMhelix
227 - 249
outside
250 - 263
TMhelix
264 - 282
inside
283 - 288
TMhelix
289 - 311
outside
312 - 320
TMhelix
321 - 343
inside
344 - 349
TMhelix
350 - 372
outside
373 - 386
TMhelix
387 - 409
inside
410 - 438
Population Genetic Test Statistics
Pi
188.802508
Theta
152.925481
Tajima's D
0.90046
CLR
0.200035
CSRT
0.630368481575921
Interpretation
Uncertain
