Gene
KWMTBOMO12021
Pre Gene Modal
BGIBMGA004355
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_ras-responsive_element-binding_protein_1_[Bombyx_mori]
Transcription factor
Location in the cell
Nuclear Reliability : 4.686
Sequence
CDS
ATGAGGAATCGTACGACTAGTATTGACAGCGACAACCGTTCGGATAGGAGCGACGATGAGGACGTCAGAAGGGCGCGAAAGCGGGCCGCGTCCACTTCTCCTTCCCCCATCCCGATGGACAAGAGGGTCGCTCGCTTCGACTGCCCGAAGTGCCGCCAGCGTTTCGATTCGGTCAACGCTTTCGACATTCACCGGTTCACAGCGCACGGAGATGATTCGAGAACGGGCCACAACGAGGTCAGTCTCGTCGACTACTCGACAAAGAAGATTCCAGAAATTATACGTGCCAGGAACCACGTCTCATCGACCGAACAACGATACAAATGTGACGTGTGCTCGAGGGATCTCCCCAATATTCAAGTATTTGAGATTCACAGGAAAACATGCACACCGAATCGTGGCTTGAGTCCGATACGCGACAGACGAGAAGAATTCTTGGCTAATCTAGATCTGCGGACCAAATCGTTCGGGATTCCGGGCACGTTGACGCCGCCGATGGAGAGATTTACATCGAAGTTCGAAGACGGGCATCTCGCGACTAACGGTATTAGACATATCGATGCAGCTCGAGATTTGGCAGATATTCAATCGATATTGAACGTCACGTCTGCGGGGAGTCTTCTCGAGAGACTCACTGGTAACAGAGTAGCTTTAGAGAGCACTGTACTTACGCCTCCAGACACCGTCGTTAAAGAACGAGAGACTGAAGAGACACAGGATAACTTTGCCGCTGAATTCAGGAGGATGAAGCTTCGGGGAGAGTTCCCTTGTCGCTTGTGTCCAGCTAAATTTCCCAATCTTAGAGCTTTAAAGGGCCATAACAGAGTCCACTTAAGTGGAACAGGACCGGGCCCTTATCAGTGCAACATGTGTCCACACGCATCTTTAGATAAGGCGGCTCTAGTGAGACATATGAGGACACATAACGGTGATCGTCCATATGAGTGTGCGGTGTGTAATTACGCGTTCACTACGAAAGCAAACTGCGAGAGACATTTAAGAAACAGACACGCCAAAGTAACCCGTGAAGATGTAAAACGCTCCATCATTTACCATCCGTCCGAAGATCCGAATAACGACGAAGTCAACTCGAAGCTCGCACGCGATGAAGTCAAGAGATCTCTCGCGTTCCACACCCCAGATATGGATAGGCGAAATGAGAGCACCGGTCGCGTGACTCCGCTAACTCATTTTACGCCTAGTTTCATTGCCGAAAGACATCCGGTCACAACACTTACATCGAAAGCTTTACCGGAAGCGCCGCTTTTGCGCGAATCAGACCGTCCCACGCCAAGGATCAAAGTGAAAGGAATCGGTCAGTTAAATCAAATCCCTGAATTCAGATCACCAGAACAAAATTACAAAACCAATGACGCTCATTCCGAAACCAAAAAGAAACGCGATGCCGACGATGAACGCAAAACCCCTGTACGCCCTACATTCGAACCAGATCCTGCGTCCGTTGCTGCTTCGGCATTAGAAAAAACGAGACTACTCTTAGCCCAGCAAAGATTATTTGAAACGTCGCTCCCAAAAATTGACCCAGCTTACTATGCTACTCATCTATCTCAAATATACGCTGGTGCGGTGCCAGGAATTCCTGGGCTACCTCTGCCCCCGTCTTTTCCTTACTTTCTTCAACCCTCGTTTTTCCCCCACCCCTCTGACACACAAGAACTTGCAGAGATAAAGCAACGTATTCAAAAAGAAATCATCCGTGGGCTGAGTATGTCCGGTGGAAGGCTTATTCCAAATGAACAAGAAACTGTTCCTAAAGAAGAACCCGAGGAAGAAGAAGTGAAGCCAATTCCACAAATGTCATCGCCTGCGCTGTCGCCTCACCCAGAATCTCCCCGACCCCTGAGCAATAACCTCGTCCCTCAGACAGACTCCGTAAAGATGGTCATTAAAAATGGGGTGCTTATGCCAAAACAGAAACAAAGACGCTATCGTACAGAAAGACCATTTTCTTGCTCACAGTGCTCTGCTAGATTCACGTTGAGATCTAACATGGAAAGGCATGTAAAACAACAGCATCCCCAACACTGGAGTGTCCGTAGACCCGCTCAAAGAGGCCCCCCACCTTACACCTCTACGGACGCTTTGGCCGATCGCGTTAAATTCGCCCTTTTGGCTCATCACCTTGAACGACCACTGCAGTCAGAGAGATCGCCGGCGCGACGAGACTCTGATGAAGTCGCCGATAATGAAGAAGATGAGGACGACGCCCTAATAATTGACGAGGAAACGGATGTGGAAACCAAGTCCGAAGAACATACTGCAGCCCGACAAGCAGCTGCCGAAATACTGATGGCTTCGCGACAACAAGAGATCAATAAAGACTTTGATCTTAAAATAGCAGGCAATTTAATTAATAAACCAGTTCCCATAGCAGAGAAAACAGAATCTGGACCCGACCCTACACCAGGACAAGATTTGCTGACCGTTCCAGTTGTACCCACACGCAGTGACGAGGAAGAAGACGAAGAAGGCCTGGTAGCTTCAACAAGCGAGGGAAATAACTCGGGCAGCGACGAAAATAAATCTGAATCTGATGTCGCACAACCACCGAAAAAGAAATCGGCGTACAGTCTCGCACCGAATAGGGTCTCGTGCCCTTACTGCCATAGAAAGTTCCCGTGGTCTTCGTCCCTCCGCCGTCACGTCCTCACTCACACGGGCCAGAAGCCGTTCAAGTGTCCCCACTGCCCTCTACTTTTCACTACCAAATCGAATTGTGACCGTCATCTGCTAAGAAAACATGGGGGGTCAGCGCGTGCCATACTCGCCGAGCCCATACCGGACACCGTGATGGCTCCCCAGCCCGCGAATGAGAACAGGAGTGTTCCAGAGAGGCCTTTCAAATGTGCATCATGCCCAACCAGCACGTTCTCCTCTATAGAAACTTTAAAGAAGCACATGTCCTCTCGGCACGGGACCGACTCGCGGCCGAGCTCGCCAAACCTGGAGATTATTGACGACAACTCTGAGATGGGCCTCGCTTTCAAGTGTCATTTGTGCGAGGCGTCCTTCGGCGACAGAAGCGGGGCCCTCGCGCATTTGGCAGCCACACATGCGACTGAATACGAACAGCTGGTGAATAAGGGGGCGTTGGATGCGGCCAGCGACAGGAGTGAAAGCGCTGACGATGACGACAGGGGCAAGTTTCCCGATCACGCTAATCGAAAGCAAATCGTTATCTAA
Protein
MRNRTTSIDSDNRSDRSDDEDVRRARKRAASTSPSPIPMDKRVARFDCPKCRQRFDSVNAFDIHRFTAHGDDSRTGHNEVSLVDYSTKKIPEIIRARNHVSSTEQRYKCDVCSRDLPNIQVFEIHRKTCTPNRGLSPIRDRREEFLANLDLRTKSFGIPGTLTPPMERFTSKFEDGHLATNGIRHIDAARDLADIQSILNVTSAGSLLERLTGNRVALESTVLTPPDTVVKERETEETQDNFAAEFRRMKLRGEFPCRLCPAKFPNLRALKGHNRVHLSGTGPGPYQCNMCPHASLDKAALVRHMRTHNGDRPYECAVCNYAFTTKANCERHLRNRHAKVTREDVKRSIIYHPSEDPNNDEVNSKLARDEVKRSLAFHTPDMDRRNESTGRVTPLTHFTPSFIAERHPVTTLTSKALPEAPLLRESDRPTPRIKVKGIGQLNQIPEFRSPEQNYKTNDAHSETKKKRDADDERKTPVRPTFEPDPASVAASALEKTRLLLAQQRLFETSLPKIDPAYYATHLSQIYAGAVPGIPGLPLPPSFPYFLQPSFFPHPSDTQELAEIKQRIQKEIIRGLSMSGGRLIPNEQETVPKEEPEEEEVKPIPQMSSPALSPHPESPRPLSNNLVPQTDSVKMVIKNGVLMPKQKQRRYRTERPFSCSQCSARFTLRSNMERHVKQQHPQHWSVRRPAQRGPPPYTSTDALADRVKFALLAHHLERPLQSERSPARRDSDEVADNEEDEDDALIIDEETDVETKSEEHTAARQAAAEILMASRQQEINKDFDLKIAGNLINKPVPIAEKTESGPDPTPGQDLLTVPVVPTRSDEEEDEEGLVASTSEGNNSGSDENKSESDVAQPPKKKSAYSLAPNRVSCPYCHRKFPWSSSLRRHVLTHTGQKPFKCPHCPLLFTTKSNCDRHLLRKHGGSARAILAEPIPDTVMAPQPANENRSVPERPFKCASCPTSTFSSIETLKKHMSSRHGTDSRPSSPNLEIIDDNSEMGLAFKCHLCEASFGDRSGALAHLAATHATEYEQLVNKGALDAASDRSESADDDDRGKFPDHANRKQIVI
Summary
Uniprot
EMBL
BABH01022158
BABH01022159
NWSH01001038
PCG72975.1
KQ459460
KPJ00857.1
+ More
GDQN01006241 JAT84813.1 AGBW02008312 OWR53764.1 ODYU01002580 SOQ40270.1 KQ461181 KPJ07682.1 GDQN01007943 JAT83111.1 JTDY01000540 KOB76731.1 GEZM01017182 JAV90849.1 KQ971352 EFA06638.2 UFQS01000668 UFQT01000668 SSX05968.1 SSX26325.1 LJIG01002743 KRT84197.1 AJVK01030638 AJVK01030639 AJVK01030640 GFDF01009631 JAV04453.1 GFDF01009632 JAV04452.1 GFDF01009630 JAV04454.1 AJWK01002093 AJWK01002094 KQ976885 KYN07528.1 KQ982021 KYN30568.1 KQ976419 KYM89288.1 GL441034 EFN65181.1
GDQN01006241 JAT84813.1 AGBW02008312 OWR53764.1 ODYU01002580 SOQ40270.1 KQ461181 KPJ07682.1 GDQN01007943 JAT83111.1 JTDY01000540 KOB76731.1 GEZM01017182 JAV90849.1 KQ971352 EFA06638.2 UFQS01000668 UFQT01000668 SSX05968.1 SSX26325.1 LJIG01002743 KRT84197.1 AJVK01030638 AJVK01030639 AJVK01030640 GFDF01009631 JAV04453.1 GFDF01009632 JAV04452.1 GFDF01009630 JAV04454.1 AJWK01002093 AJWK01002094 KQ976885 KYN07528.1 KQ982021 KYN30568.1 KQ976419 KYM89288.1 GL441034 EFN65181.1
Proteomes
PRIDE
Pfam
PF00096 zf-C2H2
SUPFAM
SSF57667
SSF57667
ProteinModelPortal
PDB
5YEG
E-value=2.67612e-12,
Score=178
Ontologies
GO
Topology
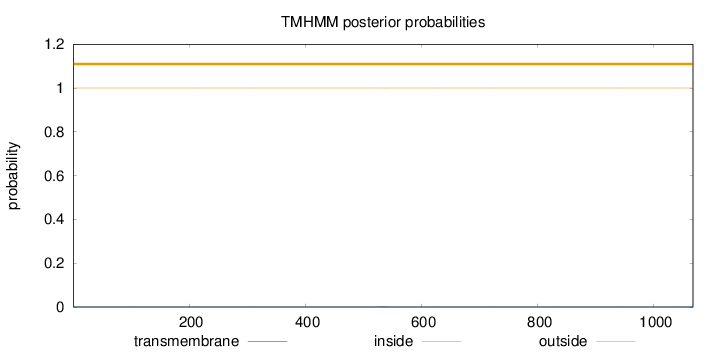
Length:
1067
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00384
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00020
outside
1 - 1067
Population Genetic Test Statistics
Pi
200.475653
Theta
178.171615
Tajima's D
0.402068
CLR
0.191157
CSRT
0.481025948702565
Interpretation
Uncertain
