Gene
KWMTBOMO12012
Pre Gene Modal
BGIBMGA004358
Annotation
PREDICTED:_focal_adhesion_kinase_1_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 2.182 Nuclear Reliability : 1.875
Sequence
CDS
ATGCAGCCCGCTCCGACGGGCGGCTCGCCCAAGCGACATGCACCCATTCAGACGCACGGCGCTGTAGCGGATCAGTCGACCTTGAAGGTGCATCTGCCGAATGGCGGCTTCAATCTGGTGCGAGCCTCGCCTGATGAAGACGTGAGGTCGGTGCTGCGACTCGTTGCCTCGAGATTAGCCTCAGGAGAGAGGGTCTACGCGTCCTGTTTTGCTTTACGAGCGAGACTCTCAAACGGGAAGATCCGATGGATTCATCAGGACACTCCCGTTTCGGAGTTGTTGACGAAGTGGCCGGCCAGCGAGTGGCGGCTCGAGCTACGAGTACGGTACCTGCCCGCGAACCTCAGGGAGCTCTGCGAAGCCGATCGCGTTACCTTCCACTATTATTACGATCAGGTGCGACACGAATACCTCAATGCCAATCATCCTACCGTAGACCAGGAGCTGGCCGTGCAAATATGTTGTTTAGAAATTAAGTACTTTTGTAAAGACATGCAGATCTCTATGGACAAAAAGTCGAACATCGAATATCTGGAGAAGGAGTTCGGCCTGCACAAGTGCTGA
Protein
MQPAPTGGSPKRHAPIQTHGAVADQSTLKVHLPNGGFNLVRASPDEDVRSVLRLVASRLASGERVYASCFALRARLSNGKIRWIHQDTPVSELLTKWPASEWRLELRVRYLPANLRELCEADRVTFHYYYDQVRHEYLNANHPTVDQELAVQICCLEIKYFCKDMQISMDKKSNIEYLEKEFGLHKC
Summary
Similarity
Belongs to the protein kinase superfamily. Tyr protein kinase family.
Uniprot
A0A212FAB3
A0A2A4IZB3
A0A2H1VD97
A0A194QQA5
A0A194QFF4
A0A2W1BP60
+ More
A0A2J7RP41 A0A0C9Q9I1 A0A0C9R9H6 A0A2J7RP27 A0A0C9RQQ4 A0A2J7RP39 A0A0C9RR47 A0A067R8A5 A0A1Y1N9J7 A0A1Y1N5U9 A0A1Y1N5Z8 A0A232EXM7 A0A195FUN6 A0A158NYB0 A0A151IX42 A0A151WIK7 E9JCV7 A0A0J7LAY4 E2AKA8 A0A026W508 A0A3L8DXL9 A0A0L7RDW5 A0A088AN13 A0A2A3EE32 A0A154PBB2 A0A310SHX3 A0A0M8ZYC2 A0A0L7LV20 A0A1B6DTP2 A0A1B6LGV8 A0A1B6LWH8 A0A1B6D3T2 A0A1B6E257 A0A1B6DL08 E2BTJ2 A0A139WEU8 D6WQS4 A0A0T6BD26 A0A1B6IZC4 A0A2G8LI83 A0A1S3GYK4 A0A1B6GEL6 V3ZMB0 T1IYC0 A0A2C9JHX6 A0A2C9JHV3 A0A2C9JHV5 A0A2C9JHW3 A0A2C9JHV8 A0A2C9JHW6 A0A2T7NP87 A0A226F2U6 A0A0B7BAE7 A0A0B7B9S0 A0A0B7B9S6 A0A2S2Q285 K1PKI5 J9JPR2 Q7Z1D3 W4YV37 T1E1S3 A0A210R109 U4U150 C3Y1A6 R7TYU7 A0A2P6KVD9 A0A1D1VCM1 N6TQ25 A0A224YYY1 A0A3R7M2I2 A0A1L8DKY7 A0A1L8DL69 A0A0L8GYB8 A0A0L8GY73 A0A131YUV2 A0A131YWN0 B9UPG7 L7MI74 A0A2R5LIB2 A0A293MH51 A0A293LJS2 A0A0P4VHC4 A0A1V9XHT5 T1I867 A0A0V0G5T5 A0A069DZP2 A0A224X6E0 A0A0A9XYV5 A0A0K8SAY9 A0A0K8S9G3
A0A2J7RP41 A0A0C9Q9I1 A0A0C9R9H6 A0A2J7RP27 A0A0C9RQQ4 A0A2J7RP39 A0A0C9RR47 A0A067R8A5 A0A1Y1N9J7 A0A1Y1N5U9 A0A1Y1N5Z8 A0A232EXM7 A0A195FUN6 A0A158NYB0 A0A151IX42 A0A151WIK7 E9JCV7 A0A0J7LAY4 E2AKA8 A0A026W508 A0A3L8DXL9 A0A0L7RDW5 A0A088AN13 A0A2A3EE32 A0A154PBB2 A0A310SHX3 A0A0M8ZYC2 A0A0L7LV20 A0A1B6DTP2 A0A1B6LGV8 A0A1B6LWH8 A0A1B6D3T2 A0A1B6E257 A0A1B6DL08 E2BTJ2 A0A139WEU8 D6WQS4 A0A0T6BD26 A0A1B6IZC4 A0A2G8LI83 A0A1S3GYK4 A0A1B6GEL6 V3ZMB0 T1IYC0 A0A2C9JHX6 A0A2C9JHV3 A0A2C9JHV5 A0A2C9JHW3 A0A2C9JHV8 A0A2C9JHW6 A0A2T7NP87 A0A226F2U6 A0A0B7BAE7 A0A0B7B9S0 A0A0B7B9S6 A0A2S2Q285 K1PKI5 J9JPR2 Q7Z1D3 W4YV37 T1E1S3 A0A210R109 U4U150 C3Y1A6 R7TYU7 A0A2P6KVD9 A0A1D1VCM1 N6TQ25 A0A224YYY1 A0A3R7M2I2 A0A1L8DKY7 A0A1L8DL69 A0A0L8GYB8 A0A0L8GY73 A0A131YUV2 A0A131YWN0 B9UPG7 L7MI74 A0A2R5LIB2 A0A293MH51 A0A293LJS2 A0A0P4VHC4 A0A1V9XHT5 T1I867 A0A0V0G5T5 A0A069DZP2 A0A224X6E0 A0A0A9XYV5 A0A0K8SAY9 A0A0K8S9G3
Pubmed
EMBL
AGBW02009503
OWR50672.1
NWSH01004533
PCG64979.1
ODYU01001925
SOQ38808.1
+ More
KQ461181 KPJ07688.1 KQ459053 KPJ04144.1 KZ150043 PZC74530.1 NEVH01002141 PNF42593.1 GBYB01011033 JAG80800.1 GBYB01009577 JAG79344.1 PNF42590.1 GBYB01009576 JAG79343.1 PNF42592.1 GBYB01011020 JAG80787.1 KK852816 KDR15787.1 GEZM01011987 GEZM01011985 JAV93315.1 GEZM01011988 JAV93312.1 GEZM01011986 JAV93314.1 NNAY01001731 OXU23095.1 KQ981261 KYN44163.1 ADTU01003596 ADTU01003597 ADTU01003598 ADTU01003599 ADTU01003600 ADTU01003601 ADTU01003602 ADTU01003603 ADTU01003604 ADTU01003605 KQ980834 KYN12384.1 KQ983089 KYQ47664.1 GL771866 EFZ09187.1 LBMM01000095 KMR04988.1 GL440210 EFN66133.1 KK107419 EZA51098.1 QOIP01000003 RLU24689.1 KQ414613 KOC69043.1 KZ288269 PBC30043.1 KQ434864 KZC09131.1 KQ759870 OAD62388.1 KQ435824 KOX72079.1 JTDY01000033 KOB79285.1 GEDC01008289 JAS29009.1 GEBQ01017109 JAT22868.1 GEBQ01011951 JAT28026.1 GEDC01016954 JAS20344.1 GEDC01005309 JAS31989.1 GEDC01010922 JAS26376.1 GL450410 EFN81020.1 KQ971354 KYB26419.1 EFA06030.2 LJIG01001741 KRT85199.1 GECU01015433 JAS92273.1 MRZV01000069 PIK59964.1 GECZ01008909 JAS60860.1 KB202050 ESO92503.1 JH431679 PZQS01000010 PVD22981.1 LNIX01000001 OXA63808.1 HACG01042167 HACG01042168 HACG01042170 CEK89032.1 CEK89033.1 CEK89035.1 HACG01042164 HACG01042166 CEK89029.1 CEK89031.1 HACG01042165 HACG01042169 HACG01042171 CEK89030.1 CEK89034.1 CEK89036.1 GGMS01002672 MBY71875.1 JH818685 EKC24552.1 ABLF02029484 AY144487 AAN38839.1 AAGJ04170363 AAGJ04170364 AAGJ04170365 AAGJ04170366 AAGJ04170367 AAGJ04170368 AAGJ04170369 AAGJ04170370 AAGJ04170371 AAGJ04170372 GAKT01000030 JAA93032.1 NEDP02000979 OWF54581.1 KB631982 ERL87594.1 GG666480 EEN65646.1 AMQN01001956 KB307198 ELT99103.1 MWRG01004713 PRD30297.1 BDGG01000004 GAU98565.1 APGK01025701 KB740600 ENN80113.1 GFPF01007824 MAA18970.1 QCYY01002485 ROT69985.1 GFDF01007047 JAV07037.1 GFDF01006994 JAV07090.1 KQ419936 KOF81917.1 KOF81918.1 GEDV01006235 JAP82322.1 GEDV01006236 JAP82321.1 EU927416 ACL14651.1 GACK01002061 JAA62973.1 GGLE01005147 MBY09273.1 GFWV01020299 MAA45027.1 GFWV01008998 MAA33727.1 GDKW01002760 JAI53835.1 MNPL01010758 OQR72953.1 ACPB03011989 ACPB03011990 GECL01003076 JAP03048.1 GBGD01000295 JAC88594.1 GFTR01008410 JAW08016.1 GBHO01019567 GBHO01019566 GBHO01019565 JAG24037.1 JAG24038.1 JAG24039.1 GBRD01015906 JAG49920.1 GBRD01015905 JAG49921.1
KQ461181 KPJ07688.1 KQ459053 KPJ04144.1 KZ150043 PZC74530.1 NEVH01002141 PNF42593.1 GBYB01011033 JAG80800.1 GBYB01009577 JAG79344.1 PNF42590.1 GBYB01009576 JAG79343.1 PNF42592.1 GBYB01011020 JAG80787.1 KK852816 KDR15787.1 GEZM01011987 GEZM01011985 JAV93315.1 GEZM01011988 JAV93312.1 GEZM01011986 JAV93314.1 NNAY01001731 OXU23095.1 KQ981261 KYN44163.1 ADTU01003596 ADTU01003597 ADTU01003598 ADTU01003599 ADTU01003600 ADTU01003601 ADTU01003602 ADTU01003603 ADTU01003604 ADTU01003605 KQ980834 KYN12384.1 KQ983089 KYQ47664.1 GL771866 EFZ09187.1 LBMM01000095 KMR04988.1 GL440210 EFN66133.1 KK107419 EZA51098.1 QOIP01000003 RLU24689.1 KQ414613 KOC69043.1 KZ288269 PBC30043.1 KQ434864 KZC09131.1 KQ759870 OAD62388.1 KQ435824 KOX72079.1 JTDY01000033 KOB79285.1 GEDC01008289 JAS29009.1 GEBQ01017109 JAT22868.1 GEBQ01011951 JAT28026.1 GEDC01016954 JAS20344.1 GEDC01005309 JAS31989.1 GEDC01010922 JAS26376.1 GL450410 EFN81020.1 KQ971354 KYB26419.1 EFA06030.2 LJIG01001741 KRT85199.1 GECU01015433 JAS92273.1 MRZV01000069 PIK59964.1 GECZ01008909 JAS60860.1 KB202050 ESO92503.1 JH431679 PZQS01000010 PVD22981.1 LNIX01000001 OXA63808.1 HACG01042167 HACG01042168 HACG01042170 CEK89032.1 CEK89033.1 CEK89035.1 HACG01042164 HACG01042166 CEK89029.1 CEK89031.1 HACG01042165 HACG01042169 HACG01042171 CEK89030.1 CEK89034.1 CEK89036.1 GGMS01002672 MBY71875.1 JH818685 EKC24552.1 ABLF02029484 AY144487 AAN38839.1 AAGJ04170363 AAGJ04170364 AAGJ04170365 AAGJ04170366 AAGJ04170367 AAGJ04170368 AAGJ04170369 AAGJ04170370 AAGJ04170371 AAGJ04170372 GAKT01000030 JAA93032.1 NEDP02000979 OWF54581.1 KB631982 ERL87594.1 GG666480 EEN65646.1 AMQN01001956 KB307198 ELT99103.1 MWRG01004713 PRD30297.1 BDGG01000004 GAU98565.1 APGK01025701 KB740600 ENN80113.1 GFPF01007824 MAA18970.1 QCYY01002485 ROT69985.1 GFDF01007047 JAV07037.1 GFDF01006994 JAV07090.1 KQ419936 KOF81917.1 KOF81918.1 GEDV01006235 JAP82322.1 GEDV01006236 JAP82321.1 EU927416 ACL14651.1 GACK01002061 JAA62973.1 GGLE01005147 MBY09273.1 GFWV01020299 MAA45027.1 GFWV01008998 MAA33727.1 GDKW01002760 JAI53835.1 MNPL01010758 OQR72953.1 ACPB03011989 ACPB03011990 GECL01003076 JAP03048.1 GBGD01000295 JAC88594.1 GFTR01008410 JAW08016.1 GBHO01019567 GBHO01019566 GBHO01019565 JAG24037.1 JAG24038.1 JAG24039.1 GBRD01015906 JAG49920.1 GBRD01015905 JAG49921.1
Proteomes
UP000007151
UP000218220
UP000053240
UP000053268
UP000235965
UP000027135
+ More
UP000215335 UP000078541 UP000005205 UP000078492 UP000075809 UP000036403 UP000000311 UP000053097 UP000279307 UP000053825 UP000005203 UP000242457 UP000076502 UP000053105 UP000037510 UP000008237 UP000007266 UP000230750 UP000085678 UP000030746 UP000076420 UP000245119 UP000198287 UP000005408 UP000007819 UP000007110 UP000242188 UP000030742 UP000001554 UP000014760 UP000186922 UP000019118 UP000283509 UP000053454 UP000192247 UP000015103
UP000215335 UP000078541 UP000005205 UP000078492 UP000075809 UP000036403 UP000000311 UP000053097 UP000279307 UP000053825 UP000005203 UP000242457 UP000076502 UP000053105 UP000037510 UP000008237 UP000007266 UP000230750 UP000085678 UP000030746 UP000076420 UP000245119 UP000198287 UP000005408 UP000007819 UP000007110 UP000242188 UP000030742 UP000001554 UP000014760 UP000186922 UP000019118 UP000283509 UP000053454 UP000192247 UP000015103
Interpro
IPR001245
Ser-Thr/Tyr_kinase_cat_dom
+ More
IPR000299 FERM_domain
IPR029071 Ubiquitin-like_domsf
IPR011009 Kinase-like_dom_sf
IPR019748 FERM_central
IPR011993 PH-like_dom_sf
IPR041390 FADK_N
IPR035963 FERM_2
IPR041784 FAK1/PYK2_FERM_C
IPR008266 Tyr_kinase_AS
IPR020635 Tyr_kinase_cat_dom
IPR005189 Focal_adhesion_kin_target_dom
IPR036137 Focal_adhe_kin_target_dom_sf
IPR019749 Band_41_domain
IPR000719 Prot_kinase_dom
IPR014352 FERM/acyl-CoA-bd_prot_sf
IPR017441 Protein_kinase_ATP_BS
IPR001412 aa-tRNA-synth_I_CS
IPR000299 FERM_domain
IPR029071 Ubiquitin-like_domsf
IPR011009 Kinase-like_dom_sf
IPR019748 FERM_central
IPR011993 PH-like_dom_sf
IPR041390 FADK_N
IPR035963 FERM_2
IPR041784 FAK1/PYK2_FERM_C
IPR008266 Tyr_kinase_AS
IPR020635 Tyr_kinase_cat_dom
IPR005189 Focal_adhesion_kin_target_dom
IPR036137 Focal_adhe_kin_target_dom_sf
IPR019749 Band_41_domain
IPR000719 Prot_kinase_dom
IPR014352 FERM/acyl-CoA-bd_prot_sf
IPR017441 Protein_kinase_ATP_BS
IPR001412 aa-tRNA-synth_I_CS
Gene 3D
ProteinModelPortal
A0A212FAB3
A0A2A4IZB3
A0A2H1VD97
A0A194QQA5
A0A194QFF4
A0A2W1BP60
+ More
A0A2J7RP41 A0A0C9Q9I1 A0A0C9R9H6 A0A2J7RP27 A0A0C9RQQ4 A0A2J7RP39 A0A0C9RR47 A0A067R8A5 A0A1Y1N9J7 A0A1Y1N5U9 A0A1Y1N5Z8 A0A232EXM7 A0A195FUN6 A0A158NYB0 A0A151IX42 A0A151WIK7 E9JCV7 A0A0J7LAY4 E2AKA8 A0A026W508 A0A3L8DXL9 A0A0L7RDW5 A0A088AN13 A0A2A3EE32 A0A154PBB2 A0A310SHX3 A0A0M8ZYC2 A0A0L7LV20 A0A1B6DTP2 A0A1B6LGV8 A0A1B6LWH8 A0A1B6D3T2 A0A1B6E257 A0A1B6DL08 E2BTJ2 A0A139WEU8 D6WQS4 A0A0T6BD26 A0A1B6IZC4 A0A2G8LI83 A0A1S3GYK4 A0A1B6GEL6 V3ZMB0 T1IYC0 A0A2C9JHX6 A0A2C9JHV3 A0A2C9JHV5 A0A2C9JHW3 A0A2C9JHV8 A0A2C9JHW6 A0A2T7NP87 A0A226F2U6 A0A0B7BAE7 A0A0B7B9S0 A0A0B7B9S6 A0A2S2Q285 K1PKI5 J9JPR2 Q7Z1D3 W4YV37 T1E1S3 A0A210R109 U4U150 C3Y1A6 R7TYU7 A0A2P6KVD9 A0A1D1VCM1 N6TQ25 A0A224YYY1 A0A3R7M2I2 A0A1L8DKY7 A0A1L8DL69 A0A0L8GYB8 A0A0L8GY73 A0A131YUV2 A0A131YWN0 B9UPG7 L7MI74 A0A2R5LIB2 A0A293MH51 A0A293LJS2 A0A0P4VHC4 A0A1V9XHT5 T1I867 A0A0V0G5T5 A0A069DZP2 A0A224X6E0 A0A0A9XYV5 A0A0K8SAY9 A0A0K8S9G3
A0A2J7RP41 A0A0C9Q9I1 A0A0C9R9H6 A0A2J7RP27 A0A0C9RQQ4 A0A2J7RP39 A0A0C9RR47 A0A067R8A5 A0A1Y1N9J7 A0A1Y1N5U9 A0A1Y1N5Z8 A0A232EXM7 A0A195FUN6 A0A158NYB0 A0A151IX42 A0A151WIK7 E9JCV7 A0A0J7LAY4 E2AKA8 A0A026W508 A0A3L8DXL9 A0A0L7RDW5 A0A088AN13 A0A2A3EE32 A0A154PBB2 A0A310SHX3 A0A0M8ZYC2 A0A0L7LV20 A0A1B6DTP2 A0A1B6LGV8 A0A1B6LWH8 A0A1B6D3T2 A0A1B6E257 A0A1B6DL08 E2BTJ2 A0A139WEU8 D6WQS4 A0A0T6BD26 A0A1B6IZC4 A0A2G8LI83 A0A1S3GYK4 A0A1B6GEL6 V3ZMB0 T1IYC0 A0A2C9JHX6 A0A2C9JHV3 A0A2C9JHV5 A0A2C9JHW3 A0A2C9JHV8 A0A2C9JHW6 A0A2T7NP87 A0A226F2U6 A0A0B7BAE7 A0A0B7B9S0 A0A0B7B9S6 A0A2S2Q285 K1PKI5 J9JPR2 Q7Z1D3 W4YV37 T1E1S3 A0A210R109 U4U150 C3Y1A6 R7TYU7 A0A2P6KVD9 A0A1D1VCM1 N6TQ25 A0A224YYY1 A0A3R7M2I2 A0A1L8DKY7 A0A1L8DL69 A0A0L8GYB8 A0A0L8GY73 A0A131YUV2 A0A131YWN0 B9UPG7 L7MI74 A0A2R5LIB2 A0A293MH51 A0A293LJS2 A0A0P4VHC4 A0A1V9XHT5 T1I867 A0A0V0G5T5 A0A069DZP2 A0A224X6E0 A0A0A9XYV5 A0A0K8SAY9 A0A0K8S9G3
PDB
4EKU
E-value=4.80084e-23,
Score=262
Ontologies
GO
Topology
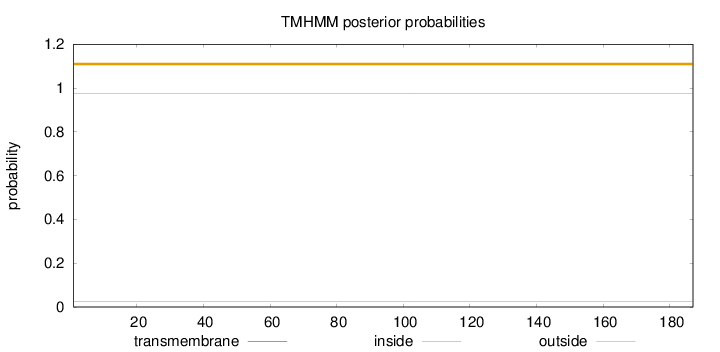
Length:
187
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00116
Exp number, first 60 AAs:
0.00032
Total prob of N-in:
0.02393
outside
1 - 187
Population Genetic Test Statistics
Pi
246.448897
Theta
154.786229
Tajima's D
2.028537
CLR
0.180091
CSRT
0.887405629718514
Interpretation
Uncertain
